Abstract
Adenosine-to-inosine (A-to-I) RNA editing, constituting nearly 90% of all RNA editing events in humans, has been reported to contribute to the tumorigenesis in diverse cancers. However, the comprehensive map for functional A-to-I RNA editing events in cancers is still insufficient. To fill this gap, we systematically and intensively analyzed multiple tumorigenic mechanisms of A-to-I RNA editing events in samples across 33 cancer types from The Cancer Genome Atlas. For individual candidate among ∼ 1,500,000 quantified RNA editing events, we performed diverse types of downstream functional annotations. Finally, we identified 24,236 potentially functional A-to-I RNA editing events, including the cases in APOL1, IGFBP3, GRIA2, BLCAP, and miR-589-3p. These events might play crucial roles in the scenarios of tumorigenesis, due to their tumor-related editing frequencies or probable effects on altered expression profiles, protein functions, splicing patterns, and microRNA regulations of tumor genes. Our functional A-to-I RNA editing events (https://ccsm.uth.edu/CAeditome/) will help better understand the cancer pathology from the A-to-I RNA editing aspect.
Keywords: A-to-I RNA editing, Cancer, Protein recoding, Alternative splicing, MicroRNA regulation
Introduction
Adenosine-to-inosine (A-to-I) RNA editing is the most common RNA editing type in humans, constituting nearly 90% of all RNA editing events. Recently, increasing evidence has revealed a significant contribution of RNA editing to tumorigenesis through multiple mechanisms [1], [2], including alteration of protein-coding capacity, generation of diverse protein isoforms, and change of cellular fate of RNA and its likelihood of being translated. Specifically, A-to-I RNA editing in coding sequences can result in the functional alterations of proteins that have roles in tumors. For example, an A-to-I RNA editing of SLC22A3, resulting in the substitution of asparagine 72 to aspartate, drives early tumor invasion and metastasis in familial esophageal cancer [3]. A study of gastric cancer has reported that editing at codon 241 of PODXL confers a loss-of-function phenotype that neutralizes the tumorigenic ability of the unedited gene [4]. Also, A-to-I RNA editing can modulate splicing to generate diverse isoforms associated with cancer. In acute myeloid leukemia, an experiment in vitro provided the evidence of aberrant intron-retaining splice variant caused by the hyper-editing of PTPN6, which is potentially involved in leukemogenesis [5]. STAT3β, the tumor regression-associated isoform, is preferentially induced by an A-to-I RNA editing event residing in proximity to the alternatively spliced exon [6]. Besides, microRNAs (miRNAs) and the three prime untranslated regions (3′-UTRs) of mRNAs can also undergo A-to-I RNA editing, which may affect their interactions in cancer. For example, an RNA editing site in miR-200b has been reported to switch the functional roles of this miRNA in terms of cell migration and invasion from suppression to promotion [7]. The edited mature miR-455-5 caused the reduction of tumor growth and metastasis by promoting tumor suppressor gene (TSG) CPEB1 in melanoma [8]. As shown in these examples, the systematic and intensive analyses of A-to-I RNA editing will provide critical evidence and novel therapeutic targets in human cancers.
To date, there are several pan-cancer editing landscapes covering the functional annotations of RNA editing events from the aspects of clinical associations [9], [10], [11], protein recoding [9], and miRNA regulations [7], [11]. For these aspects, they either provided limited candidates for each cancer type, or included partial analyses of RNA editing. For a more comprehensive map of functional A-to-I RNA editing events, in this study, we performed a systematic and intensive bioinformatics analysis pipeline (Figure 1) for all the samples across 33 cancer types from The Cancer Genome Atlas (TCGA), similar as that was used in the data analyses of Alzheimer’s disease from our recent study [12]. All the analyses were cooperative to point out 24,236 functional A-to-I RNA editing candidates and present their potential roles in the scenarios of tumorigenesis. From the analyses, we confirmed the possible functions of the well-known R/G editing (GRIA2, CAediting_390714) in neurological and brain tumors, expanded the roles of BLCAP Q/R editing (CAediting_1426931) in carcinogenesis promotion in pan-cancers, and re-addressed the tumorigenic control potential of edited miR-589-3p (CAediting_524911) through dysregulations of tumor genes (TGs). In addition, we also studied two another novel and promising functional RNA editing events. One case (CAediting_1478179) was up-edited in diverse cancers and may confer its pathological function through the intervention in miRNA regulation on the TG of APOL1. Another event (CAediting_543208) occurred only in tumor samples for multiple cancer types and may enhance the ability of IGFBP3 to inhibit tumor cell growth. All these discoveries are available at https://ccsm.uth.edu/CAeditome/. This database provides novel knowledge of tumorigenesis and lists potential targets for cancer and drug research communities.
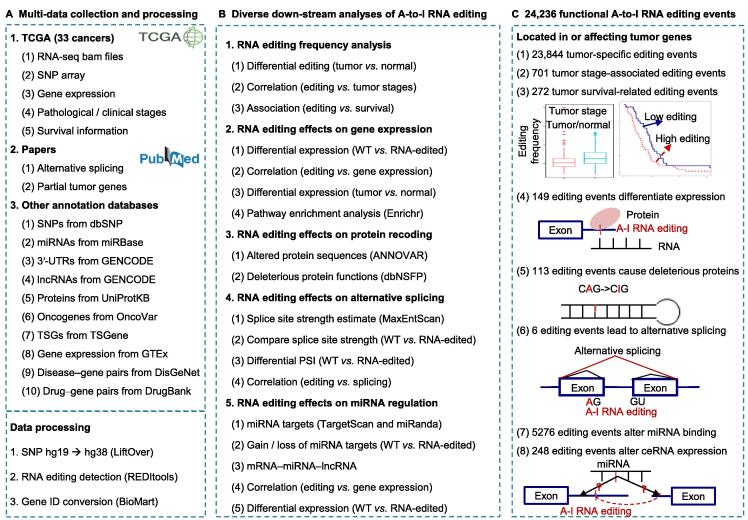
Figure 1.
The flowchart to identify functional A-to-I RNA editing events in cancers
A. The collection and pre-processing of multi-omics data across 33 cancer types. B. Diverse down-stream analyses of A-to-I RNA editing events. C. The potentially functional A-to-I RNA editing events related to tumorigenesis. A-to-I, adenosine-to-inosine; TCGA, The Cancer Genome Atlas; RNA-seq, RNA sequencing; SNP, single-nucleotide polymorphism; miRNA, microRNA; 3′-UTR, 3′-untranslated region; lncRNA, long non-coding RNA; TSG, tumor suppressor gene; GTEx, Genotype-Tissue Expression; WT, wild-type.
Results
RNA editing candidates are abnormally edited in cancers
The changes of editing frequencies in tumors, along with diverse stages of tumor pathology and across different tumor survival statuses, can reveal aberrant RNA editing events probably responsible for tumor occurrence, progression, and poor survival. In this work, after comparing editing frequencies between tumor samples and controls, we identified 23,844 RNA editing events in 869 TGs showing tumor-specific frequencies (Figure 2A–F, Figures S1–S4; Table S1). Next, through the correlation studies of editing frequencies with tumor stages, we found 701 RNA editing events in 158 TGs which were significantly associated with tumor progression (Figure 2G–I, Figures S5 and S6; Table S1). Then the survival analysis discovered 272 RNA editing events in 99 TGs which might affect the survival risks of cancer patients (Figure 2J–L, Figures S7 and S8; Table S1). Among the 23,904 functional RNA editing events, we selected two candidates to show the effects of A-to-I RNA editing on tumors.
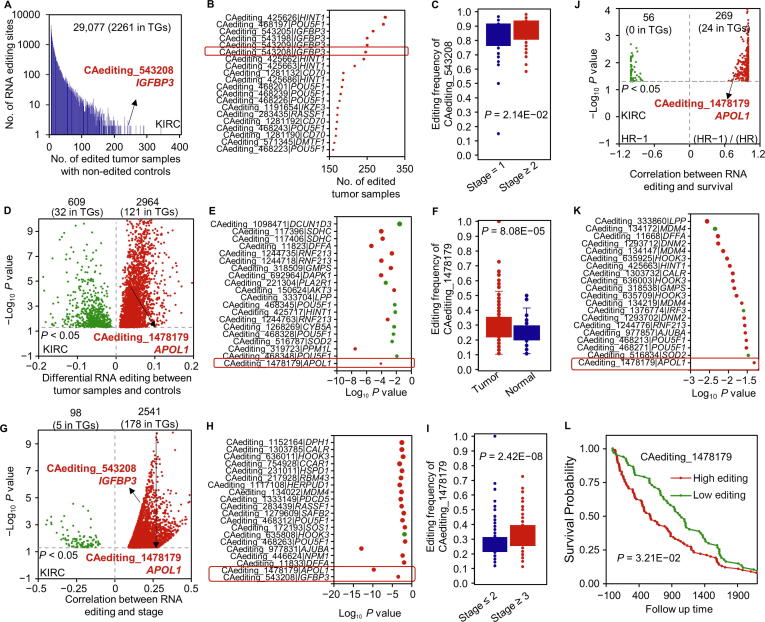
Figure 2.
RNA editing frequency analysis
A. KIRC-specific A-to-I RNA editing events with more than five edited tumor samples and non-edited normal controls. The histogram presents the distribution of this kind of RNA editing events along with the number of edited tumor samples. B. The bubble plot introduces a part of KIRC-specific RNA editing events in TGs which are not edited in normal controls. C. One significant case of IGFBP3 occurred only in 246 tumor samples for the KIRC cancer type, also showing higher editing frequencies in more severe KIRC tumors. D. KIRC-specific A-to-I RNA editing events showing differential editing frequencies in KIRC tumors compared with controls. The volcano plot presents the differences of editing frequencies between tumor samples and controls. E. The bubble plot introduces a part of KIRC-specific RNA editing events in TGs which are differentially edited in tumor samples compared with normal controls. F. One significant case in APOL1 showed higher editing frequencies in KIRC tumor samples. G. KIRC stage-associated A-to-I RNA editing events. The volcano plot presents the correlations of editing frequencies with tumor stages. H. The bubble plot introduces a part of KIRC stage-associated RNA editing events in TGs. I. One significant case in APOL1 showed higher editing frequencies in more severe KIRC tumors. J. KIRC survival-related A-to-I RNA editing events. The volcano plot presents the correlations of editing frequencies with cancer survival. K. The bubble plot introduces a part of KIRC survival-related RNA editing events in TGs. L. One significant case in APOL1 showed higher editing frequencies in the poorer survival group. The analysis results of editing frequencies for other cancer types are displayed in Figures S1–S8. KIRC, kidney renal clear cell carcinoma; TG, tumor gene; HR, hazard ratio.
One is an RNA editing event in Chr22:36266650 (CAediting_1478179) of the APOL1 gene in the cancer type of kidney renal clear cell carcinoma (KIRC). This event showed significantly higher editing frequencies in tumor samples than controls with a P value of 8.08E−05 (Figure 2F), as well as higher editing frequencies in more severe tumor samples (t-test: P = 2.42E−08; Spearman test: P = 1.71E−10 and R = 0.28) (Figure 2I) and a high risk of KIRC survival [Kaplan–Meier (KM) analysis: P = 3.21E−02; Cox proportional–hazards regression (COX) analysis: P = 4.75E−02 and hazard ratio (HR) = 3.42] (Figure 2L). Its existence in the 3′-UTR of APOL1 seems to cause the up-regulation of the edited gene (t-test: P = 1.04E−14 and log2 fold change (FC) = 1.86; Pearson test: P = 7.44E−06 and R = 0.20) (Figure 3A and B) from the loss of original miR-7151-3p binding targets detected by TargetScan [13] and miRanda [14]. Due to the up-regulated expression of this gene in KIRC tumor samples compared with controls (P = 6.42E−21 and log2 FC = 2.04) (Figure 3C), and its inducing role in autophagy [15], we suggest this RNA editing event as a potential biomarker of KIRC progression and survival (Figure 3D).
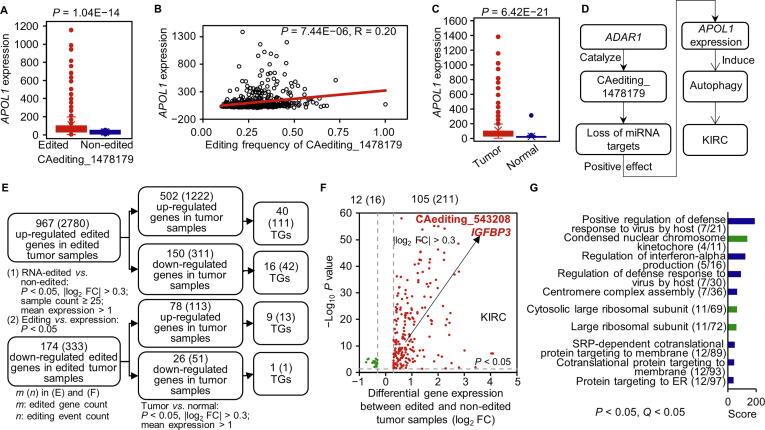
Figure 3.
The effects of A-to-I RNA editing events on gene expression
A.APOL1 was up-regulated in the RNA-edited group. B. The expression levels of APOL1 were positively associated with the frequencies of the CAediting_1478179 editing event. C.APOL1 was abnormally expressed in KIRC tumor samples compared with controls. D. The CAediting_1478179 editing event seems to be a potential biomarker for the KIRC cancer type, because it caused the loss of original miRNA binding targets to induce the up-regulated expression of APOL1, which may interfere in the autophagy function of this gene in cancer. E. The analysis procedure for the effects of A-to-I RNA editing events on gene expression. First, we performed a DEG analysis between RNA-edited and non-edited tumor samples, as well as a correlation analysis between gene expression and editing frequency to identify the DEGs whose expression levels were probably affected by A-to-I RNA editing. Then, we overlapped these genes with the DEGs identified in tumor samples compared to normal controls, to focus on RNA editing effects on the aberrantly expressed genes in cancers, especially the TGs. F. The overlapping DEGs in the KIRC cancer type. G. The overlapping DEGs were enriched in the immune- and replication-related functions and processes. The RNA editing effects on gene expression in other cancer types are presented in Figures S10–S13. FC, fold change; DEG, differentially expressed gene.
Another editing candidate locates in Chr7:45916046 (CAediting_543208) of the IGFBP3 gene as shown in Figure S9. For the KIRC cancer type, this event occurred only in tumor samples (246/535 vs. 0/72) and was up-edited in the samples with higher stages (t-test: P = 2.14E−02; Spearman test: P = 3.04E−04 and R = 0.23) (Figure 2C). Moreover, it seems to be linked with the up-regulated expression of IGFBP3 (t-test: P = 2.18E−09 and log2 FC = 0.61; Pearson test: P = 2.06E−02 and R = 0.15). Because this gene was up-regulated in tumor samples (P = 1.81E−90 and log2 FC = 3.47), and seemed to act in an autocrine action to suppress tumor cell growth [16], we may carefully suggest the role of this RNA editing event in enhancing the protective functions of IGFBP3 against cancer progression.
RNA editing candidates are potential factors to affect TG expression
From the analyses mentioned above, we found that the frequencies of several RNA editing events were significantly associated with the expression of their host genes involved in tumors. For systematic analysis of the potential contributions of these RNA editing events to the expression levels of the edited genes, we performed a differentially expressed gene (DEG) analysis between RNA-edited and non-edited tumor samples, as well as a correlation analysis between gene expression and editing frequency (Figures S10 and S11; Table S1). Then, the A-to-I RNA editing-affected DEGs that overlapped with the DEGs between tumor samples and controls were used for following enrichment analysis, to understand the probably involved pathways and biological functions of these RNA editing events in cancers.
As shown in Figure 3E–F, we first discovered 2780 and 333 RNA editing events that would cause the up- and down-regulated expression of 967 and 174 edited genes, respectively. Of them, 651 genes were also abnormally expressed in tumor samples compared with controls, including 55 TGs possibly affected by 149 A-to-I RNA editing events. Combining the potential promotion or inhibition roles of these edited genes in cancers, we could infer the possible functions of these RNA editing events related to tumorigenesis, such as CAediting_1478179 of APOL1 and CAediting_543208 of IGFBP3 mentioned above.
Besides, Figure S12 presents a well-known R/G editing case in Chr4:157360142 position (CAediting_390714) of GRIA2 (syn. GluA2) in the cancer type of pheochromocytoma and paraganglioma (PCPG). Its editing frequency was positively associated with the expression of GRIA2 (t-test: P = 5.04E−13 and log2 FC = 1.02; Pearson test: P = 1.34E−02 and R = 0.20). Due to the up-regulated expression of this gene in tumors (P = 4.64E−23 and log2 FC = 5.11), and its possible roles in proliferation stimulation, apoptosis resistance, migration, and invasion in cancer cell lines [17], this editing event in GRIA2 may be a pathological biomarker for the PCPG cancer type, which was also supported by 146 edited PCPG tumor samples and non-edited normal samples.
The following enrichment analysis for these 651 edited DEGs revealed immune- and replication-related biological functions and processes (Figure 3G). Specifically, activation of innate and adaptive immune response through the regulation of viral defense and interferon-alpha production is benefit for cancer immunotherapy [18], [19]. The replication processes related to ribosome, endoplasmic reticulum, kinetochore, and so on are important for tumor proliferation and cancer risks [20], [21], [22]. Moreover, for each cancer type, the enrichment analysis also discovered some tumor-related Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways as shown in Figure S13 and Table S2. For example, targeting apoptosis is a promising therapy to eliminate cancer cells [23], antigen processing and presentation pathway (APP) is the cellular mechanism that determines direct interactions between cancer cells and adaptive immune system [24], and sphingolipids metabolic network provides regulatory nodes for controlling tumor growth and proliferation in response to cellular stress [25]. Then, the DEG-associated RNA editing events probably affect these pathways or processes involved in cancers.
RNA editing candidates may reshape their protein functions in tumorigenesis
RNA editing events in coding regions can alter amino acid sequences and have a chance to affect protein functions. To study this, we first selected A-to-I RNA editing sites in protein-coding sequences and identified 3785 non-synonymous and 121 stop-loss editing events (Figure 4A). Out of these, 1128 RNA editing sites were recognized to have impacts on the biological functions of 491 proteins by at least one of the annotation tools such as SIFT, Polyphen2, and PROVEAN. Among them, 113 A-to-I RNA editing events may reshape the functions of 52 tumor-related proteins (Figure 4B).
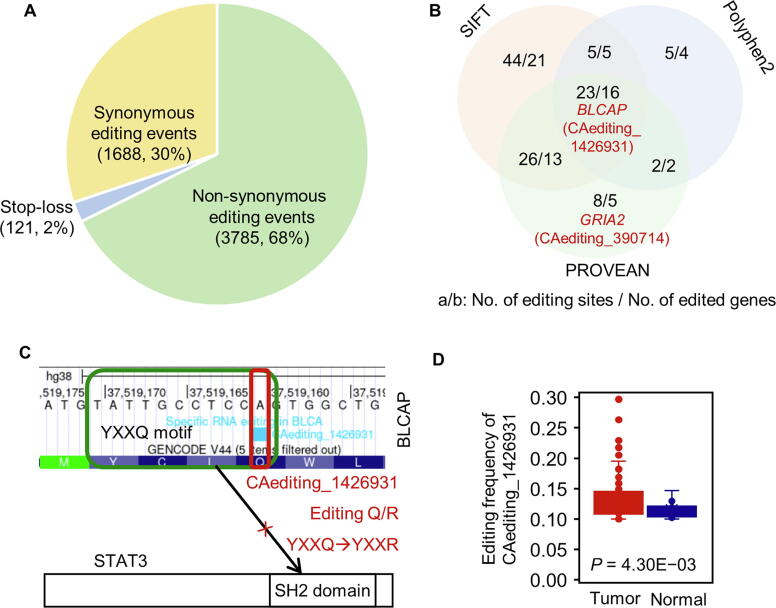
Figure 4.
The effects of A-to-I RNA editing events on protein recoding
A. There are 3785 non-synonymous and 121 stop-loss editing events causing the changes of amino acid sequences. B. 113 A-to-I RNA editing events conferred their deleterious effects on 52 tumor-related proteins assessed by SIFT, Polyphen2, and PROVEAN. Among these, there were 12 proteins with different RNA editing events whose effects were predicted to be diverse by these three tools. C. The Q/R editing in the key YXXQ motif of the BLCAP protein reverses the inhibition ability of BLCAP to STAT3, potentially facilitating the cancer-initiating and progressing events. D. The hypothesis in (C) was supported by its higher editing frequency in breast invasive carcinoma.
One RNA editing candidate in the position of Chr20:37519161 (CAediting_1426931) leads to the Q/R changes of key YXXQ motif in the BLCAP protein (Figure 4C). This editing event reverses the inhibition ability of BLCAP to STAT3, facilitating the cancer-initiating and progressing events [26]. Its roles in carcinogenesis promotion were also supported by (1) its abnormal editing cases in pan-cancers, such as the higher editing frequencies in the cancer types of breast invasive carcinoma (BRCA; P = 4.30E−03) (Figure 4D) and KIRC (P = 4.33E−02), (2) positive associations with tumor stages for bladder urothelial carcinoma (BLCA; P = 3.03E−02 and R = 0.29), and (3) mere occurrence in tumor samples of BLCA (55/411 vs. 0/19), colon adenocarcinoma (COAD; 176/471 vs. 0/41), head and neck squamous cell carcinoma (HNSC; 9/501 vs. 0/44), cholangiocarcinoma (CHOL; 10/36 vs. 0/9), and rectum adenocarcinoma (READ; 70/167 vs. 0/10).
Another well-known R/G editing event (CAediting_390714) in the coding region of the GRIA2 protein mediates the fast excitatory synaptic transmission [27] and may affect the functions of this gene in tumor cell growth, migration, and invasion [17]. Its roles in cancers were also supported by its differential editing frequencies in glioblastoma multiforme (GBM; P = 1.78E−02) (Figure S3). Therefore, the other 111 RNA editing events may also possibly be involved in cancers through modifying the functions of tumor-related proteins, which deserve to be studied further.
RNA editing candidates are probable regulators of alternative splicing in TGs
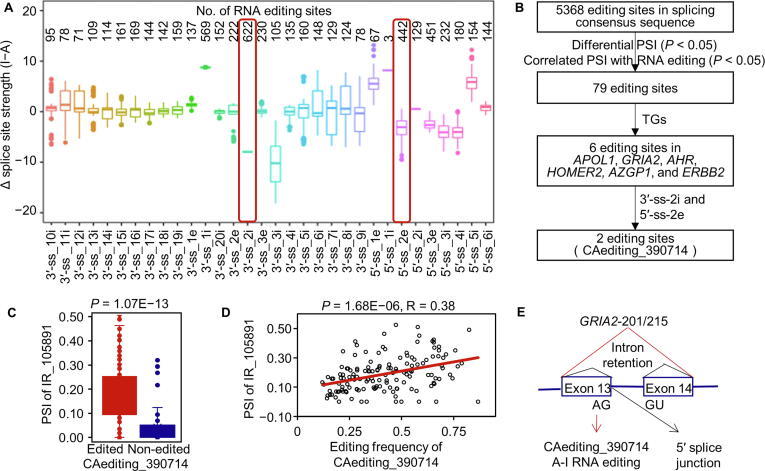
RNA editing sites in the alternatively spliced exon regions can differentiate splice site strength and eventually affect the selection of splicing positions. To study this, we focused on the editing sites locating around the exon junction boundaries for all the 33 cancer types. In total, we identified 3600 RNA editing sites in the 3′-acceptor splice site (3′-ss) regions and 1779 RNA editing sites in the 5′-donor splice site (5′-ss) regions. They present diverse impacts on the splice site strength of 1957 genes, due to their different locations in the splicing sequences (Figure 5A). Among these editing events, 79 cases have verified their effects on alternative splicing (Figure 5B), through the differential percent spliced in (PSI) values in RNA-edited samples and significant correlations of PSI values with editing frequencies. Out of them, 6 A-to-I RNA editing events may have an opportunity to be involved in autophagy reduction [15], tumor growth [28], cell proliferation [17], [29], cancer metastasis [17], [29], toxicity mediation [30], and so on, because they altered the splicing patterns of TGs.
Figure 5.
The effects of A-to-I RNA editing events on alternative splicing
A. The distribution of altered splice site strength caused by RNA editing events in the different positions of splicing regions. Individual RNA editing site may belong to different groups according to different exons. B. The analysis procedure for the effects of A-to-I RNA editing events on alternative splicing. C. The intron retention event (IR_105891) was mostly occurred in the RNA-edited group of PCPG. D. The intron retention event (IR_105891) was associated with the frequency of the editing event (CAediting_390714) in PCPG. E. A hypothesis that the R/G editing in GRIA2 may alter the canonical splicing pattern of AG-GU to induce the intron retention for the isoforms of tumor-related GRIA2 in PCPG. 3′-ss, 3′-acceptor splice site; 5′-ss, 5′-donor splice site; PSI, percent spliced in; PCPG, pheochromocytoma and paraganglioma.
One case of them is the well-known R/G editing (CAediting_390714) in GRIA2. It altered the canonical splicing pattern of AG-GU and caused a reduction of the 5′-donor splice site strength (5.37 − 8.23 = −2.86) to potentially induce the intron retention (157360143:157361009) for the isoforms of GRIA2 in the PCPG cancer type (Figure 5C–E). According to all the analyses mentioned above, this editing event may play important roles in neurological or brain cancers through altering the excitatory synaptic transmission by its contributions to protein function changes, alternative splicing, and dysregulation of GRIA2 (Figure S12).
RNA editing candidates likely intervene in miRNA regulation on TGs
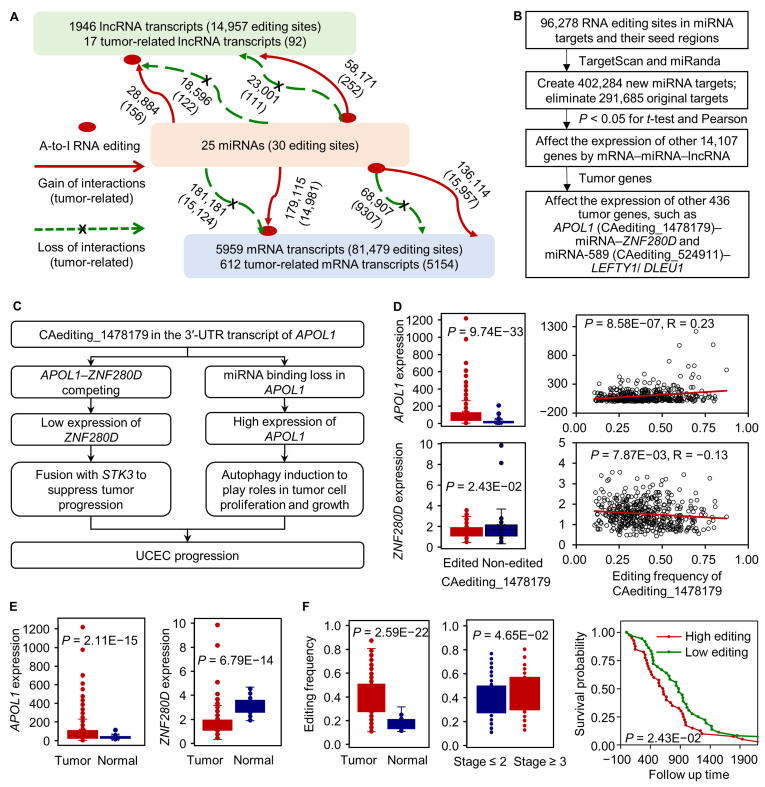
RNA editing sites in the binding targets of miRNAs or their seed regions would alter miRNA–RNA interactions and possibly affect the expression of miRNA-regulated genes. Through miRNA target prediction for wild-type (WT) and RNA-edited transcripts, we identified 96,278 RNA editing sites in 7930 transcripts, which were presumed to potentially create 402,284 new miRNA targets and eliminate 291,685 original ones (Figure 6A). These altered miRNA–target interactions further conferred their effects on the expression of 14,107 regulated genes (Table S1) other than the edited genes. Taking into account the functions of all these genes in tumor, we eventually selected 248 functional A-to-I RNA editing candidates, which may likely intervene in miRNA regulation on 436 TGs to play their roles in tumorigenesis (Figure 6B).
Figure 6.
The effects of A-to-I RNA editing events on miRNA regulation
A. RNA editing events in the 3′-UTRs of mRNAs, lncRNAs, and miRNA seed regions led to the changes of miRNA–target interactions. Individual RNA editing site may locate in both of protein-coding or non-coding transcripts due to their possible overlaps. B. The analysis procedure for the effects of A-to-I RNA editing events on miRNA regulation. C. One significant RNA editing event (CAediting_1478179) in the 3′-UTR of APOL1 likely intervened in the miRNA regulation on two TGs (APOL1 and ZNF280D) to play its roles in the UCEC progression. D. The RNA editing event (CAediting_1478179) caused the loss of original miRNA binding target on APOL1. The altered miRNA regulation resulted in the increased expression of APOL1 and indirectly led to the reduced expression of competing ZNF280D gene. E.APOL1 and ZNF280D showed differential expression in UCEC, revealing their potential roles and functions in UCEC. F. Analyses uncovered CAediting_1478179 in the 3′-UTR of APOL1 as a probably pathological biomarker of UCEC. Another significant case shown in panel B is described in Figure S14. UCEC, uterine corpus endometrial carcinoma.
For example, an RNA editing site (CAediting_1478179) in the 3′-UTR of APOL1-201/202/205/206 isoforms would lead to the loss of original binding targets of miR-7151-3p (Figure 6C). The lost regulation seems to cause the increased expression of APOL1 and indirectly lead to the reduced expression of ZNF280D with long non-coding RNA (lncRNA) transcripts in uterine corpus endometrial carcinoma (UCEC), due to their competing relationships (Figure 6D). For these two genes, several recent studies reported the induction function of APOL1 in autophagy [15] to probably promote tumor cell growth and proliferation [31], and the fusion possibility of ZNF280D with the TSG of STK3 [32], [33] to involve in cancer. Besides, the DEG analysis also revealed the up-regulation of APOL1 and down-regulation of ZNF280D in the UCEC tumor samples (Figure 6E). Therefore, we may infer CAediting_1478179 as a progression biomarker for the UCEC cancer type. It was also supported by the significantly higher frequencies of this editing event in the tumor samples, along with more severe tumor statuses and poorer survival probability (Figure 6F).
Another RNA editing example located in Chr7:5495852 (CAediting_524911) of miR-589-3p. It altered the miRNA binding target from original DLEU1 to LEFTY1 (Figure S14). The lost miRNA regulation led to the up-regulated expression of DLEU1, whereas the gained interactions caused the down-regulated expression of LEFTY1 in the RNA-edited testicular germ cell tumor (TGCT) samples. From previous studies, we found that DLEU1 is one lncRNA produced from the 13q14.3 tumor suppressor locus and regulates the NF-kB signaling pathway, which plays crucial roles in cancer initiation and progression [34], [35]. We thus inferred a tumor suppressor role of DLEU1 in TGCT because of its location in tumor suppressor locus, functions related to cancer, and also down-regulated expression in TGCT and more severe tumor samples. In addition, LEFTY1, a key gene in the Nodal pathway, was reported to be specifically associated with germ cell pluripotency, the presence of carcinoma in situ, and TGCT [36]. Moreover, this gene was also verified to be up-regulated in TCGT and along with more severe tumor samples. Due to the functions and expression alterations of these two genes, we could speculate that the edited miR-589-3p might potentially alleviate TGCT tumor condition, through the up-regulation of DLEU1 and down-regulation of LEFTY1. In conclusion, these two editing candidates represent the potential functions of A-to-I RNA editing events in cancers through altering the miRNA regulation on TGs.
Discussion
Through measuring and analyzing RNA editing events in human pan-cancers, we could provide 24,236 potentially functional A-to-I RNA editing candidates (Table S3). They either were abnormally edited in cancers or might alter the original expression profiles, protein functions, splicing patterns, or miRNA regulation of TGs. Considering the contributions of these TGs to glutamine metabolism [17], modified immunity [26], selective autophagy [15], DNA damage responses [37], and so on, we may infer that the functional A-to-I RNA editing events may play important roles in tumorigenesis. In the future, the appearance of more functional RNA editing candidates will be accompanied by the increase of TGs. The possible RNA editing events and their functions were all archived in CAeditome database (https://ccsm.uth.edu/CAeditome/).
Among them, five events were explored in detail to introduce their potential functions related to cancers in this study. They are CAediting_390714 of GRIA2, CAediting_1426931 of BLCAP, CAediting_1478179 of APOL1, CAediting_543208 of IGFBP3, and CAediting_524911 of miR-589-3p. The R/G editing in GRIA2 (CAediting_390714) was studied deeply in previous work for its involvements in the desensitization of AMPA receptor (AMPAR) channels [27], AMPAR-mediated neurotransmission, and neurodevelopmental deficits [38], [39]. For its potentials in cancers, one previous study validated its roles in tumor survival, cell viability, and targeted therapeutics [9]. In our study, for this editing event, we discovered its anomalously lower editing frequencies in GBM, and positive associations with aberrant expression profiles and alternative splicing values of GRIA2 in PCPG. Due to the roles of this gene in proliferation stimulation, apoptosis resistance, migration, and invasion in cancer cell lines [17], we may suggest the possible bi-functions of this RNA editing event in neurological and brain tumors. As for the Q/R editing in BLCAP (CAediting_1426931), it was abnormally edited in multiple cancer types, such as the mere occurrence in the tumor samples of BLCA, COAD, HNSC, CHOL, and READ, higher editing frequencies in BRCA and KIRC, and positive associations with BLCA tumor stages. The analyses in our study expanded its roles of carcinogenesis promotion in pan-cancers from the cervical cancer reported in previous literature [26]. The third RNA editing event (CAediting_1478179) seems to be a novel and promising pathological biomarker for various cancer types including cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), COAD, and esophageal carcinoma (ESCA). Especially, it displayed remarkably up-regulated editing frequencies in tumors, more severe tumor samples, and poorer survival groups for the cancer types of KIRC (tumor vs. normal: P = 8.08E−05; editing vs. stage: P = 1.71E−10 and R = 0.28; editing vs. survival: PKM = 3.21E−02, PCOX = 4.75E−02, and HR = 3.42), lung adenocarcinoma (LUAD) (tumor vs. normal: P = 1.52E−25; editing vs. stage: P = 6.59E−03 and R = 0.12; editing vs. survival: PKM = 2.36E−02, PCOX = 6.81E−03, and HR = 3.88), and UCEC (tumor vs. normal: P = 2.59E−22; editing vs. stage: P = 4.32E−02 and R = 0.10; editing vs. survival: PKM = 2.43E−02, PCOX = 1.50E−02, and HR = 5.74). Moreover, we discovered that it caused the loss of original miRNA binding targets to potentially induce the up-regulated expression of APOL1, which was supported by their positive associations in pan-cancers, such as for KIRC (P = 7.44E−06 and R = 0.20), LUAD (P = 6.78E−10 and R = 0.27), and UCEC (P = 8.58E−07 and R = 0.23). Thus, this RNA editing event may confer its pathological function in cancers through its intervention in miRNA regulation on the TG of APOL1. Another novel RNA editing event (CAediting_543208) occurred only in the tumor samples for multiple cancer types, especially for KIRC (246/535 vs. 0/72). It may enhance the inhibition ability of tumor cell growth through its positive impacts on the TSG of IGFBP3 [16]. The last RNA editing event (CAediting_524911) was located in the seed region of miR-589-3p to potentially modify its original regulations on many TGs. For example, the edited miR-589-3p altered the expression levels of DLEU1 and LEFTY1, which thus may alleviate TGCT tumor condition. Moreover, another evidence in one previous study also validated the regulatory potential of this RNA editing event (CAediting_524911) on two genes of PCDH9 and ADAM12 to control glioblastoma cell migration and invasion [40].
Of the five edited genes introduced in this study, three were discovered to be linked with tumor-related phenotypes from DisGeNET (January 2021, v.7.0) [41]. Specifically, APOL1 is associated with neoplasm-related nephrotic syndrome and common focal segmental glomerulosclerosis form of kidney disease, IGFBP3 plays important roles in multiple cancers, and GRIA2 is related to neurological or brain diseases. Their possible relationships with these disorders may be partially attributed to the RNA editing events in them, which may also affect the effectiveness of probable drugs targeting them. In total, we discovered 6717 edited genes associated with 9203 different types of diseases in DisGeNET, and 1586 edited genes targeted by 1674 approved drugs from DrugBank (January 2021, v.5.1.8) [42]. The functions of A-to-I RNA editing events in these genes will be useful for exploring the pathological mechanisms of related diseases and providing novel knowledge to design the targeted drugs.
For the two novel RNA editing biomarkers in APOL1 and IGFBP3, we also performed replication analyses in the lung squamous cell carcinoma (LUSC) samples from Cancer Cell Line Encyclopedia (CCLE) [43]. The analysis results (Figure S15) also supported the associations between these two editing events and their host genes mentioned in this study. Moreover, because CCLE contained metastasis information for each sample, we selected one RNA editing biomarker (CAediting_279186) in the metastasis-related gene of RHOA [44] for further validation. As shown in Figure S16, this editing event was negatively associated with RHOA expression. Given the abnormal expression of RHOA in primary tumors and metastasis samples, we could annotate the potential functions of this RNA editing event in cancer metastasis. The hypothesis was also partially held up after the analyses of this event in another metastasis dataset, MET500 [45].
Besides the five RNA editing events introduced in detail, there are also several potential candidates archived in CAeditome database that were validated or proposed in previous studies. For example, in our study, the analysis of RNA editing events in protein-coding regions identified I164V in COPA (CAediting_115738), S367G in AZIN1 (CAediting_655260), and I635V in COG3 (CAediting_962851). These three RNA editing events were also abnormally edited in tumor samples, correlated with tumor severity, associated with cancer survival, and possible factors to affect the expression of their host genes in multiple cancer types. Their functions in tumorigenesis have been validated in cell lines and mice models by other groups [9], [46], [47], [48]. In addition, the analysis of RNA editing events in miRNA seed regions uncovered altered miR-200b regulation associated with CAediting_393. This event leads to the gain or loss of many miRNA binding targets, including that in LIFR, ZEB1, and ZEB2. Although the expression of these three genes was not significantly associated with the frequencies of this editing event as shown in different cell lines previously [7], [49], we still included it as a potentially functional A-to-I RNA editing candidate, because it probably dysregulated other tumor-related genes (Download Page in CAeditome database). For example, ACSL6 was down-regulated to possibly interfere in the metabolites of fatty acids, abnormality of which is one of the cancer hallmarks [50]. Another gene PKP1 was inhibited to prevent the survival and metastasis of cancer cells by decreasing cluster formation in circulatory system [51]. Moreover, there are also other RNA editing events whose functions in cancers were bio-experimentally validated, including H241R editing (CAediting_604339) in PODXL [4], K242R/K242E (CAediting_1062027/CAediting_1062028) editing in NEIL1 [52], and two editing events (CAediting_442015/CAediting_442019) in the 3′-UTR of GM2A [53]. The other potential RNA editing biomarkers are waiting for the validation by cancer research communities.
In summary, this study proposed a transcriptome-wide and cancer-wide map for the functions of individual A-to-I RNA editing events. It will provide the chances to understand cancer pathology from the A-to-I RNA editing aspect and list potential biomarkers and therapeutic targets for cancer and drug research communities. However, during the analyses, the complex regulatory mechanisms associated with RNA editing pointed out two possible studies in the future.
Firstly, we noted the possible interactions of multiple RNA editing events and their co-effects on the downstream genes or regulations. For example, three RNA editing events were all in together to confer their effects on the expression of APOL1, which were deciphered by the least absolute shrinkage and selection operator regression method as shown in Figure S17. In the future, to uncover the co-regulation of RNA editing events, we will propose an RNA editing weighted gene expression network and evaluate its usefulness in various clinical scenarios such as survival prognosis.
On the other hand, we confirmed the deamination functions of three editing enzymes on 11,948 RNA editing events in this study (P < 0.05 and R > 0.3) (Figure S18). However, we should not ignore that there are another 28,062 RNA editing events showing no statistical associations with all the three enzymes. It revealed the possible multi-regulators of A-to-I RNA editing. From previous literature, these diverse regulatory mechanisms probably include genetic variations [54], splicing efficiency [55], and RNA binding proteins [56]. For a functional RNA editing candidate (CAediting_1478179 of APOL1) proposed in this study, we also discovered its differential editing frequencies among the genotyping groups of three single-nucleotide polymorphisms (SNPs) in the KIRC cancer type (Figure S19). For the impacts of genetic variants on RNA editing events, recently, Leng Han group has published a database named GPEdit [57]. The studies on the other potential regulators of A-to-I RNA editing are our further research plans.
Materials and methods
Detection of A-to-I RNA editing
For all the 11,056 RNA sequencing samples across 33 cancer types in TCGA (Table S1), we first detected RNA editing candidates by the script of REDItoolKnown.py (REDItools v.1.2.1) [58] with default settings (i.e., minimal read coverage = 10; minimal quality score = 30; and minimal mapping quality score = 255) and the hg38 reference files (GENCODE v.22) same as that used in the Genomic Data Commons (GDC) data harmonization and generation pipelines. These detected candidates were then checked for their reliability. Only the candidates, which occurred in the REDIportal database (January 2021) [59] but did not belong to SNPs (dbSNP151 and Genome-Wide Human SNP Array 6.0 in hg38 version converted by LiftOver [60]), were covered by more than three edited reads, and showed editing frequencies higher than 0.1, were considered as reliable editing sites. Eventually, we selected one kind of RNA editing types, A-to-I RNA editing, for further analysis, because of its abundance in humans.
For all the detected A-to-I RNA editing events, we analyzed their distributions in diverse genomic locations and repeats by ANNOVAR [61], and evaluated the stability alterations of edited transcripts by RNAfold (ViennaRNA v.2.4.17) [62]. The consistency of these analysis results (Figures S20 and S21) with previous publications [10], [12] revealed the reliability of A-to-I RNA editing detection in this study. Moreover, all these events covered a high ratio of RNA editing sites (72.49%) detected in one previous study [9] using a different pipeline as shown in Figure S22. It described the consistency of our RNA editing detection pipeline with others. In addition, the more RNA editing sites detected in our study provided an opportunity for a more comprehensive map of functional A-to-I RNA editing events in cancers.
Analysis of A-to-I RNA editing frequencies
To uncover potential A-to-I RNA editing events related to tumors, we first compared their editing frequencies between tumor samples and normal controls across 33 cancer types. Then we defined a tumor-specific RNA editing event if it only occurred in tumors with more than 5 edited samples or showed significantly differential editing frequency (P < 0.05) in tumor samples. Next, we analyzed the correlations between editing frequency and tumor stage (pathologic stage or clinical stage, P < 0.05), to identify tumor progression-associated RNA editing events. Third, we performed KM and COX analyses to determine the A-to-I RNA editing events (P < 0.05 for both results) that may affect tumor survival. Last, we focused on the RNA editing events in 1615 tumor-related genes, including driver oncogenes from OncoVar [63], TSGs from TSGene 2.0 [64], and some other genes reported to be associated with tumors in previous literature [15], [17], in order to further analyze the possible effects of A-to-I RNA editing in cancers.
Analysis of A-to-I RNA editing effects on gene expression and pathways
To study the effects of A-to-I RNA editing events on gene expression, we performed the analyses of DEGs between RNA-edited and non-edited tumor samples (P < 0.05 and |log2 FC| > 0.3) by t-test and Pearson correlations between editing frequency and corresponding gene expression (P < 0.05) across 33 cancer types. The dysregulated genes in RNA-edited tumor groups and also along with the changes of editing frequency were inferred to be potentially affected by A-to-I RNA editing. The genes which overlapped with the DEGs identified in tumors compared with controls (t-test: P < 0.05 and |log2 FC| > 0.3) were further studied by Enrichr [65], to assess the probably involved cellular processes of A-to-I RNA editing in cancers.
Analysis of A-to-I RNA editing effects on protein recoding and functions
For A-to-I RNA editing events in coding regions, we used ANNOVAR to detect the changes of amino acid sequences caused by the non-synonymous and stop-loss editing sites. These alterations are shown in lollipop figures with editing sites and UniProt KnowledgeBase (UniProtKB) protein IDs [66] converted from BioMart [67]. Then, their deleterious effects on protein functions were assessed by SIFT, Polyphen2, and PROVEAN (dbNSFP v.4.1a).
Analysis of A-to-I RNA editing effects on alternative splicing of pre-mRNAs
To study the effects of A-to-I RNA editing events on splicing, we first overlapped them with 5′-ss and 3′-ss regions around detected exons [68]. The 5′-ss region is a 9-mer region of 3 nt in the exon and 6 nt in the intron, whereas the 3′-ss region is a 23-mer region of 3 nt in the exon and 20 nt in the intron based on a previous splicing study [69]. MaxEntScan method proposed in that study was also used here to estimate the changes of splice site strength for sequences being edited. These splicing alterations were further validated by the comparisons of PSI values between RNA-edited and non-edited tumor samples (P < 0.05) and the correlations of PSI values with corresponding editing frequencies (P < 0.05), to discover the reliable effects of A-to-I RNA editing events on splicing patterns.
Analysis of A-to-I RNA editing effects on miRNA regulation
For the WT and RNA-edited 3′-UTRs of mRNAs, lncRNAs, and miRNA seed regions, we used TargetScan (v.7.0) and miRanda (v.3.3a) to detect miRNA binding targets. Based on the predicted miRNA–lncRNA/mRNA 3′-UTR interactions, we defined the gain of miRNA binding targets as the interactions existing in the RNA-edited sequences but not in the WT sequences supported by both tools and vice versa for the loss of miRNA binding targets. Furthermore, we checked the expression of miRNA-regulated genes between RNA-edited and non-edited tumor groups (P < 0.05) and also along with the changes of editing frequencies (P < 0.05), to discover the altered miRNA regulation caused by these A-to-I RNA editing events.
Data availability
All the analysis results involved in this study are available at https://ccsm.uth.edu/CAeditome/.
Competing interests
The authors have declared no competing interests.
CRediT authorship contribution statement
Sijia Wu: Conceptualization, Data curation, Methodology, Investigation, Software, Formal analysis, Writing – original draft, Funding acquisition. Zhiwei Fan: Visualization. Pora Kim: Conceptualization, Visualization, Validation, Writing – review & editing. Liyu Huang: Supervision, Project administration, Resources, Funding acquisition. Xiaobo Zhou: Supervision, Project administration, Resources. All authors have read and approved the final manuscript.
Acknowledgments
The results here are in whole or part based upon data generated by the TCGA Research Network in https://www.cancer.gov/tcga. This work was supported by the National Natural Science Foundation of China (Grant No. 62002270), the Fundamental Research Funds for the Central Universities, the Natural Science Foundation of Shaanxi Province of China (Grant No. 2020JQ-332), the China Postdoctoral Science Foundation (Grant No. 2018M643583), the National Natural Science Foundation of China (Grant No. 82227802), and the National Key R&D Program of China (Grant No. 2017YFA0205202), and partially funded by the National Natural Science Foundation of China (Grant No. 61672422). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Handled by Leng Han
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation and Genetics Society of China.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gpb.2022.12.010.
Contributor Information
Pora Kim, Email: Pora.Kim@uth.tmc.edu.
Liyu Huang, Email: huangly@mail.xidian.edu.cn.
Xiaobo Zhou, Email: Xiaobo.Zhou@uth.tmc.edu.
Supplementary material
The following are the Supplementary data to this article:
Tumor-specific RNA editing events with more than 5 edited tumor samples and none edited normal controls The X-axis indicates the number of edited tumor samples and the Y-axis represents the number of editing sites. The genes shown in this figure are tumor-related genes with this kind of tumor-specific RNA editing events. Specifically, CAediting_543208 (Chr7:45916046) of IGFBP3 occurred only in tumor samples (246/535) and none in controls (0/72) for KIRC. BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRP, kidney renal papillary cell carcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; STAD, stomach adenocarcinoma; THCA, thyroid carcinoma.
The bubble plots of top tumor-specific RNA editing events (edited only in tumor samples with number ≥ 5) in tumor genes for each cancer type The X-axis shows the number of edited tumor samples, and the Y-axis represents the editing events and their host tumor genes
The volcano plots for tumor-specific RNA editing events (P < 0.05 and edited samples ≥ 50) The X-axis denotes the differences of editing frequencies between tumor samples and controls, and the Y-axis represents −log10P value. The genes shown in this figure are tumor-related genes with this kind of tumor-specific RNA editing events. CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; PAAD, pancreatic adenocarcinoma.
The bubble plots of top tumor-specific RNA editing events (P < 0.05 and edited samples ≥ 50) in tumor genes for each cancer type The X-axis shows log10P value, and the Y-axis represents the editing events and their host tumor genes. The size of each dot denotes the absolute differences of editing frequencies. The red and green dots represent the RNA editing events showing significantly higher or lower frequencies in tumor samples.
The volcano plots for tumor stage-associated RNA editing events (P < 0.05 and edited tumor samples ≥ 50) The X-axis denotes the correlation coefficients between editing frequencies and tumor stages, and the Y-axis represents −log10P value. The genes shown in this figure are tumor-related genes with tumor stage-associated RNA editing events. ACC, adrenocortical carcinoma; MESO, mesothelioma; OV, ovarian serous cystadenocarcinoma; SKCM, skin cutaneous melanoma; TGCT, testicular germ cell tumors; UCS, uterine carcinosarcoma; UVM, uveal melanoma.
The bubble plots of top tumor stage-associated RNA editing events (P < 0.05 and edited tumor samples ≥ 50) in tumor genes for each cancer type The X-axis shows log10P value, and the Y-axis represents the editing events and their host tumor genes. The size of each dot denotes the absolute correlation coefficients. The red and green dots represent the RNA editing events showing positive or negative relationships with tumor stages.
The volcano plots for tumor survival-related RNA editing events (P < 0.05 and edited tumor samples ≥ 50) If HR is smaller than 1.0, the X-axis denotes the value of HR−1. Otherwise, the X-axis denotes the value of (HR−1)/HR. The Y-axis represents −log10P value. The genes shown in this figure are tumor-related genes with tumor survival-related RNA editing events. HR, hazard ratio; LAML, acute myeloid leukemia; LGG, brain lower grade glioma; SARC, sarcoma; THYM, thymoma.
The bubble plots of top tumor survival-related RNA editing events (P < 0.05 and edited tumor samples ≥ 50) in tumor genes for each cancer type The X-axis shows log10P value, and the Y-axis represents the editing events and their host tumor genes. The size of each dot denotes −log10P value. The red and green dots represent the RNA editing events showing high (HR > 1) or low (HR < 1) survival risks for cancer patients.
The potential of CAediting_543208 (IGFBP3) in KIRC cancer type This event occurred only in tumor samples (246/535) and none in controls (0/72), and was up-edited in tumor samples with higher stages. It was also related to the over-expressions of IGFBP3, which was up-regulated in tumor samples and seemed to act in an autocrine action to suppress tumor cell growth. Thus, this RNA editing event might enhance the protection functions of IGFBP3 against cancer progression and be potential as a therapeutic target.
The volcano plots for DEG-associated RNA editing events in each cancer type These events were identified according to the procedures shown in Figure 3E. The X-axis denotes log2 FC value between RNA-edited and non-edited tumor samples, and the Y-axis represents −log10P value. The genes shown in this figure are tumor-related genes with DEG-associated RNA editing events.
The bubble plots of top DEG-associated RNA editing events in tumor genes for each cancer type The X-axis shows log10P value, and the Y-axis represents the editing events and their host tumor genes. The size of each dot denotes absolute log2 FC value between RNA-edited and non-edited tumor samples. The red and green dots represent the RNA editing events positively or negatively associated with the expressions of their host genes.
An example (Chr4:157360142, CAediting_390714) showing the effects of RNA editing on gene expressions The frequencies of this editing event were positively associated with the expressions of its host gene (GRIA2) in PCPG. Since this editing event led to the significantly higher expressions of its host gene, which was also up-regulated in tumor samples, CAediting_390714 may be a pathological biomarker for the PCPG cancer type. It was also supported by 146 (183) edited tumor samples and 0 (3) edited controls in this cancer type.
The enriched KEGG pathways of the overlapped DEGs for each cancer type (P < 0.05 and Q < 0.2) The pathways colored with brown are tumor-related pathways. KEGG, Kyoto Encyclopedia of Genes and Genomes.
An example showing the effects of RNA editing on miRNA regulations An RNA editing event in Chr7:5495852 (CAediting_524911) of miR589-3p altered the miRNA binding target from original DLEU1 to LEFTY1. Due to the lost regulation of miR-589-3p, DLEU1 was highly expressed in the RNA-edited TGCT tumor samples. On the other hand, LEFTY1 was down-regulated because of the gained interactions with miR-589-3p.
The possible validation of two RNA editing candidates in CCLE A. The CAediting_1478179 and CAediting_543208 were associated with its host genes in TCGA-LUSC. B. These associations were also discovered in CCLE. CCLE, Cancer Cell Line Encyclopedia.
A potential A-to-I RNA editing biomarker and its roles in cancer metastasis A. The CAediting_279186 was abnormally edited in LUSC, and also associated with RHOA expressions. B. We tested the roles of this RNA editing event between primary and lymph node metastasis lung squamous cell carcinoma samples from CCLE. C. We also validated the roles of this RNA editing between primary and metastasis lung cancer samples from MET500.
An example showing the co-effects of three RNA editing events on the expressions of APOL1 in the KIRC cancer type A. The linear regression formula to estimate the co-effects of multiple RNA editing events on gene expressions. Ck represents the contributions of the m(k-th) RNA editing event to the expression of i-th gene. B. LASSO process to select optimal parameter λ (left) and corresponding coefficients (right) for the contributions of three RNA editing events to APOL1 expressions. C. The detailed contribution coefficients are shown in this panel. LASSO, least absolute shrinkage and selection operator.
The correlations of RNA editing frequency with ADAR expression The frequencies of 9702, 2476, and 1919 RNA editing events were significantly associated with the expressions of ADAR1, ADAR2, and ADAR3, respectively (P < 0.05 and R > 0.3 for Pearson method). In addition, there are also 28,062 RNA editing events (P ≥ 0.05 for all three ADARs with more than 50 edited samples) which were possibly regulated by genetic variations, splicing efficiency, RNA binding proteins, or some other mechanisms. ADAR, adenosine deaminase action RNA.
The potentially regulatory effects of genetic variants on RNA editing A. The associations between rs2003814 and CAediting_1478179. B. The associations between rs9607326 and CAediting_1478179. C. The associations between rs9607325 and CAediting_1478179.
The distributions of A-to-I RNA editing events A. The distributions of RNA editing events in different types of genes. B. The distributions of RNA editing events in diverse regions. C. The distributions of RNA editing events in repeats. lincRNA, long intergenic non-coding RNA; ncRNA, non-coding RNA.
The effects of RNA editing on secondary structure A. An A-to-I RNA editing event will significantly reduce MFE value to stabilize RNA structure. B. 89.32% RNA transcripts were stabilized by one or multiple RNA editing events. MFE, minimum free energy.
The comparisons of RNA editing events detected by different pipelines For comparison, we converted the genome coordinates of RNA editing events detected in previous study (PMID: 26439496) from GRCh37/hg19 to GRCh38/hg38.
The statistical results for RNA editing related cancer mechanisms
The tumor-related KEGG pathways
The potentially functional A-to-I RNA editing events
References
- 1.Eisenberg E., Levanon E.Y. A-to-I RNA editing — immune protector and transcriptome diversifier. Nat Rev Genet. 2018;19:473–490. doi: 10.1038/s41576-018-0006-1. [DOI] [PubMed] [Google Scholar]
- 2.Baysal B.E., Sharma S., Hashemikhabir S., Janga S.C. RNA editing in pathogenesis of cancer. Cancer Res. 2017;77:3733–3739. doi: 10.1158/0008-5472.CAN-17-0520. [DOI] [PubMed] [Google Scholar]
- 3.Fu L., Qin Y.R., Ming X.Y., Zuo X.B., Diao Y.W., Zhang L.Y., et al. RNA editing of SLC22A3 drives early tumor invasion and metastasis in familial esophageal cancer. Proc Natl Acad Sci U S A. 2017;114:E4631–E4640. doi: 10.1073/pnas.1703178114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan T.H.M., Qamra A., Tan K.T., Guo J., Yang H., Qi L., et al. ADAR-mediated RNA editing predicts progression and prognosis of gastric cancer. Gastroenterology. 2016;151:637–650. doi: 10.1053/j.gastro.2016.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alessandro B., Ripamonti C.B., Paolo P., Gaia R., Roberto C., Enrica M., et al. RNA hyperediting and alternative splicing of hematopoietic cell phosphatase (PTPN6) gene in acute myeloid leukemia. Hum Mol Genet. 2000;9:2297–2304. doi: 10.1093/oxfordjournals.hmg.a018921. [DOI] [PubMed] [Google Scholar]
- 6.Goldberg L., Abutbul-Amitai M., Paret G., Nevo-Caspi Y. Alternative splicing of STAT3 is affected by RNA editing. DNA Cell Biol. 2017;36:367–376. doi: 10.1089/dna.2016.3575. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y., Xu X., Yu S., Jeong K.J., Zhou Z., Han L., et al. Systematic characterization of A-to-I RNA editing hotspots in microRNAs across human cancers. Genome Res. 2017;27:1112–1125. doi: 10.1101/gr.219741.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shoshan E., Mobley A.K., Braeuer R.R., Kamiya T., Huang L., Vasquez M.E., et al. Reduced adenosine-to-inosine miR-455-5p editing promotes melanoma growth and metastasis. Nat Cell Biol. 2015;17:311–321. doi: 10.1038/ncb3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han L., Diao L., Yu S., Xu X., Li J., Zhang R., et al. The genomic landscape and clinical relevance of A-to-I RNA editing in human cancers. Cancer Cell. 2015;28:515–528. doi: 10.1016/j.ccell.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chigaev M., Yu H., Samuels D.C., Sheng Q., Oyebamiji O., Ness S., et al. Genomic positional dissection of RNA editomes in tumor and normal samples. Front Genet. 2019;10:211. doi: 10.3389/fgene.2019.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin C.H., Chen S.C.C. The Cancer Editome Atlas: a resource for exploratory analysis of the adenosine-to-inosine RNA editome in cancer. Cancer Res. 2019;79:3001–3006. doi: 10.1158/0008-5472.CAN-18-3501. [DOI] [PubMed] [Google Scholar]
- 12.Wu S., Yang M., Kim P., Zhou X. ADeditome provides the genomic landscape of A-to-I RNA editing in Alzheimer’s disease. Brief Bioinform. 2021;22:bbaa384. doi: 10.1093/bib/bbaa384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agarwal V., Bell G.W., Nam J.W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Betel D., Koppal A., Agius P., Sander C., Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhaorigetu S., Wan G., Kaini R., Wan G., Jiang Z., Hu C.A. ApoL1, a BH3-only lipid-binding protein, induces autophagic cell death. Autophagy. 2008;4:1079–1082. doi: 10.4161/auto.7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheung C.W., Vesey D.A., Nicol D.L., Johnson D.W. The roles of IGF-I and IGFBP-3 in the regulation of proximal tubule, and renal cell carcinoma cell proliferation. Kidney Int. 2004;65:1272–1279. doi: 10.1111/j.1523-1755.2004.00535.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H.Y., Yang W., Lu J.B. Knockdown of GluA2 induces apoptosis in non-small-cell lung cancer A549 cells through the p53 signaling pathway. Oncol Lett. 2017;14:1005–1010. doi: 10.3892/ol.2017.6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borden E.C. Interferons α and β in cancer: therapeutic opportunities from new insights. Nat Rev Drug Discov. 2019;18:219–234. doi: 10.1038/s41573-018-0011-2. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez-Cao M., Karachaliou N., Santarpia M., Viteri S., Meyerhans A., Rosell R. Activation of viral defense signaling in cancer. Ther Adv Med Oncol. 2018;10:1758835918793105 doi: 10.1177/1758835918793105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pelletier J., Thomas G., Volarević S. Ribosome biogenesis in cancer: new players and therapeutic avenues. Nat Rev Cancer. 2018;18:51–63. doi: 10.1038/nrc.2017.104. [DOI] [PubMed] [Google Scholar]
- 21.Martinon F. Targeting endoplasmic reticulum signaling pathways in cancer. Acta Oncol. 2012;51:822–830. doi: 10.3109/0284186X.2012.689113. [DOI] [PubMed] [Google Scholar]
- 22.Zhang W., Mao J.H., Zhu W., Jain A.K., Liu K., Brown J.B., et al. Centromere and kinetochore gene misexpression predicts cancer patient survival and response to radiotherapy and chemotherapy. Nat Commun. 2016;7:12619. doi: 10.1038/ncomms12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carneiro B.A., El-Deiry W.S. Targeting apoptosis in cancer therapy. Nat Rev Clin Oncol. 2020;17:395–417. doi: 10.1038/s41571-020-0341-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mpakali A., Stratikos E. The role of antigen processing and presentation in cancer and the efficacy of immune checkpoint inhibitor immunotherapy. Cancers. 2021;13:134. doi: 10.3390/cancers13010134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogretmen B. Sphingolipid metabolism in cancer signalling and therapy. Nat Rev Cancer. 2018;18:33–50. doi: 10.1038/nrc.2017.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen W., He W., Cai H., Hu B., Zheng C., Ke X., et al. A-to-I RNA editing of BLCAP lost the inhibition to STAT3 activation in cervical cancer. Oncotarget. 2017;8:39417–39429. doi: 10.18632/oncotarget.17034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lomeli H., Mosbacher J., Melcher T., Hoger T., Geiger J.R., Kuner T., et al. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994;266:1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- 28.Luo P., Liang C., Jing W., Zhu M., Zhou H., Chai H., et al. Homer2 and Homer3 act as novel biomarkers in diagnosis of hepatitis B virus-induced hepatocellular carcinoma. J Cancer. 2021;12:3439–3447. doi: 10.7150/jca.52118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J., Han H., Fan Z., Beaino M.E., Fang Z., Li S., et al. AZGP1 inhibits soft tissue sarcoma cells invasion and migration. BMC Cancer. 2018;18:89. doi: 10.1186/s12885-017-3962-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murray I.A., Patterson A.D., Perdew G.H. Aryl hydrocarbon receptor ligands in cancer: friend and foe. Nat Rev Cancer. 2014;14:801–814. doi: 10.1038/nrc3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yun C.W., Lee S.H. The roles of autophagy in cancer. Int J Mol Sci. 2018;19:3466. doi: 10.3390/ijms19113466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim P., Zhou X. FusionGDB: fusion gene annotation DataBase. Nucleic Acids Res. 2019;47:D994–1004. doi: 10.1093/nar/gky1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X., Wang F., Zhang Z.G., Yang X.M., Zhang R. STK3 suppresses ovarian cancer progression by activating NF-κB signaling to recruit CD8+ T-cells. J Immunol Res. 2020;2020:7263602. doi: 10.1155/2020/7263602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garding A., Bhattacharya N., Claus R., Ruppel M., Tschuch C., Filarsky K., et al. Epigenetic upregulation of lncRNAs at 13q14.3 in leukemia is linked to the in cis downregulation of a gene cluster that targets NF-kB. PLoS Genet. 2013;9:e1003373. doi: 10.1371/journal.pgen.1003373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park M.H., Hong J.T. Roles of NF-κB in cancer and inflammatory diseases and their therapeutic approaches. Cells. 2016;5:15. doi: 10.3390/cells5020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spiller C.M., Bowles J., Koopman P. Nodal/Cripto signaling in fetal male germ cell development: implications for testicular germ cell tumors. Int J Dev Biol. 2013;57:211–219. doi: 10.1387/ijdb.130028pk. [DOI] [PubMed] [Google Scholar]
- 37.Chua M.W.Y., Lin M.Z., Martin J.L., Baxter R.C. Involvement of the insulin-like growth factor binding proteins in the cancer cell response to DNA damage. J Cell Commun Signal. 2015;9:167–176. doi: 10.1007/s12079-015-0262-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kubota-Sakashita M., Iwamoto K., Bundo M., Kato T. A role of ADAR2 and RNA editing of glutamate receptors in mood disorders and schizophrenia. Mol Brain. 2014;7:5. doi: 10.1186/1756-6606-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vollmar W., Gloger J., Berger E., Kortenbruck G., Köhling R., Speckmann E.J., et al. RNA editing (R/G site) and flip-flop splicing of the AMPA receptor subunit GluR2 in nervous tissue of epilepsy patients. Neurobiol Dis. 2004;15:371–379. doi: 10.1016/j.nbd.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 40.Cesarini V., Silvestris D.A., Tassinari V., Tomaselli S., Alon S., Eisenberg E., et al. ADAR2/miR-589-3p axis controls glioblastoma cell migration/invasion. Nucleic Acids Res. 2018;46:2045–2059. doi: 10.1093/nar/gkx1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piñero J., Ramírez-Anguita J.M., Saüch-Pitarch J., Ronzano F., Centeno E., Sanz F., et al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020;48:D845–D855. doi: 10.1093/nar/gkz1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghandi M., Huang F.W., Jané-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R., et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature. 2019;569:503–508. doi: 10.1038/s41586-019-1186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Struckhoff A.P., Rana M.K., Worthylake R.A. RhoA can lead the way in tumor cell invasion and metastasis. Front Biosci. 2011;16:1915–1926. doi: 10.2741/3830. [DOI] [PubMed] [Google Scholar]
- 45.Robinson D.R., Wu Y.M., Lonigro R.J., Vats P., Cobain E., Everett J., et al. Integrative clinical genomics of metastatic cancer. Nature. 2017;548:297–303. doi: 10.1038/nature23306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song Y., An O., Ren X., Chan T.H.M., Tay D.J.T., Tang S.J., et al. RNA editing mediates the functional switch of COPA in a novel mechanism of hepatocarcinogenesis. J Hepatol. 2021;74:135–147. doi: 10.1016/j.jhep.2020.07.021. [DOI] [PubMed] [Google Scholar]
- 47.Wei Y., Zhang H., Feng Q., Wang S., Shao Y., Wu J., et al. A novel mechanism for A-to-I RNA-edited AZIN1 in promoting tumor angiogenesis in colorectal cancer. Cell Death Dis. 2022;13:294. doi: 10.1038/s41419-022-04734-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng X., Xu X., Wang Y., Hawke D.H., Yu S., Han L., et al. A-to-I RNA editing contributes to proteomic diversity in cancer. Cancer Cell. 2018;33:817–828. doi: 10.1016/j.ccell.2018.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramírez-Moya J., Baker A.R., Slack F.J., Santisteban P. ADAR1-mediated RNA editing is a novel oncogenic process in thyroid cancer and regulates miR-200 activity. Oncogene. 2020;39:3738–3753. doi: 10.1038/s41388-020-1248-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sebastiano M.R., Konstantinidou G. Targeting long chain acyl-CoA synthetases for cancer therapy. Int J Mol Sci. 2019;20:3624. doi: 10.3390/ijms20153624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li K., Wu R., Zhou M., Tong H., Luo K.Q. Desmosomal proteins of DSC2 and PKP1 promote cancer cells survival and metastasis by increasing cluster formation in circulatory system. Sci Adv. 2021;7:eabg7265. doi: 10.1126/sciadv.abg7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anadón C., Guil S., Simó-Riudalbas L., Moutinho C., Setien F., Martínez-Cardús A., et al. Gene amplification-associated overexpression of the RNA editing enzyme ADAR1 enhances human lung tumorigenesis. Oncogene. 2016;35:4407–4413. doi: 10.1038/onc.2015.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang L., Hao Y., Shao C., Wu Q., Prager B.C., Gimple R.C., et al. ADAR1-mediated RNA editing links ganglioside catabolism to glioblastoma stem cell maintenance. J Clin Invest. 2022;132:e143397. doi: 10.1172/JCI143397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park E., Jiang Y., Hao L., Hui J., Xing Y. Genetic variation and microRNA targeting of A-to-I RNA editing fine tune human tissue transcriptomes. Genome Biol. 2021;22:77. doi: 10.1186/s13059-021-02287-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Licht K., Kapoor U., Amman F., Picardi E., Martin D., Bajad P., et al. A high resolution A-to-I editing map in the mouse identifies editing events controlled by pre-mRNA splicing. Genome Res. 2019;29:1453–1463. doi: 10.1101/gr.242636.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Quinones-Valdez G., Tran S.S., Jun H.I., Bahn J.H., Yang E.W., Zhan L., et al. Regulation of RNA editing by RNA-binding proteins in human cells. Commun Biol. 2019;2:19. doi: 10.1038/s42003-018-0271-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ruan H., Li Q., Liu Y., Liu Y., Lussier C., Diao L., et al. GPEdit: the genetic and pharmacogenomic landscape of A-to-I RNA editing in cancers. Nucleic Acids Res. 2022;50:D1231–D1237. doi: 10.1093/nar/gkab810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Giudice C.L., Tangaro M.A., Pesole G., Picardi E. Investigating RNA editing in deep transcriptome datasets with REDItools and REDIportal. Nat Protoc. 2020;15:1098–1131. doi: 10.1038/s41596-019-0279-7. [DOI] [PubMed] [Google Scholar]
- 59.Mansi L., Tangaro M.A., Giudice C.L., Flati T., Kopel E., Schaffer A.A., et al. REDIportal: millions of novel A-to-I RNA editing events from thousands of RNAseq experiments. Nucleic Acids Res. 2021;49:D1012–D1019. doi: 10.1093/nar/gkaa916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee B.T., Barber G.P., Benet-Pagès A., Casper J., Clawson H., Diekhans M., et al. The UCSC Genome Browser database: 2022 update. Nucleic Acids Res. 2022;50:D1115–D1122. doi: 10.1093/nar/gkab959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lorenz R., Bernhart S.H., Zu Siederdissen C.H.Z., Tafer H., Flamm C., Stadler P.F., et al. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang T, Ruan S, Zhao X, Shi X, Teng H, Zhong J, et al. OncoVar: an integrated database and analysis platform for oncogenic driver variants in cancers. Nucleic Acids Res 2021;49:D1289–301. [DOI] [PMC free article] [PubMed]
- 64.Zhao M., Pora K., Ramkrishna M., Zhao J., Zhao Z. TSGene 2.0: an updated literature-based knowledgebase for tumor suppressor genes. Nucleic Acids Res. 2016;44:D1023–D1031. doi: 10.1093/nar/gkv1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boutet E., Lieberherr D., Tognolli M., Schneider M., Bansal P., Bridge A.J., et al. UniProtKB/Swiss-Prot, the manually annotated section of the UniProt KnowledgeBase: how to use the entry view. Methods Mol Biol. 2016;1374:23–54. doi: 10.1007/978-1-4939-3167-5_2. [DOI] [PubMed] [Google Scholar]
- 67.Kinsella R.J., Kähäri A., Haider S., Zamora J., Proctor G., Spudich G., et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database. 2011;2011:bar030. doi: 10.1093/database/bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kahles A., Lehmann K.V., Toussaint N.C., Hüser M., Stark S.G., Sachsenberg T., et al. Comprehensive analysis of alternative splicing across tumors from 8705 patients. Cancer Cell. 2018;34:211–224. doi: 10.1016/j.ccell.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yeo G., Burge C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tumor-specific RNA editing events with more than 5 edited tumor samples and none edited normal controls The X-axis indicates the number of edited tumor samples and the Y-axis represents the number of editing sites. The genes shown in this figure are tumor-related genes with this kind of tumor-specific RNA editing events. Specifically, CAediting_543208 (Chr7:45916046) of IGFBP3 occurred only in tumor samples (246/535) and none in controls (0/72) for KIRC. BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRP, kidney renal papillary cell carcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; STAD, stomach adenocarcinoma; THCA, thyroid carcinoma.
The bubble plots of top tumor-specific RNA editing events (edited only in tumor samples with number ≥ 5) in tumor genes for each cancer type The X-axis shows the number of edited tumor samples, and the Y-axis represents the editing events and their host tumor genes
The volcano plots for tumor-specific RNA editing events (P < 0.05 and edited samples ≥ 50) The X-axis denotes the differences of editing frequencies between tumor samples and controls, and the Y-axis represents −log10P value. The genes shown in this figure are tumor-related genes with this kind of tumor-specific RNA editing events. CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; PAAD, pancreatic adenocarcinoma.
The bubble plots of top tumor-specific RNA editing events (P < 0.05 and edited samples ≥ 50) in tumor genes for each cancer type The X-axis shows log10P value, and the Y-axis represents the editing events and their host tumor genes. The size of each dot denotes the absolute differences of editing frequencies. The red and green dots represent the RNA editing events showing significantly higher or lower frequencies in tumor samples.
The volcano plots for tumor stage-associated RNA editing events (P < 0.05 and edited tumor samples ≥ 50) The X-axis denotes the correlation coefficients between editing frequencies and tumor stages, and the Y-axis represents −log10P value. The genes shown in this figure are tumor-related genes with tumor stage-associated RNA editing events. ACC, adrenocortical carcinoma; MESO, mesothelioma; OV, ovarian serous cystadenocarcinoma; SKCM, skin cutaneous melanoma; TGCT, testicular germ cell tumors; UCS, uterine carcinosarcoma; UVM, uveal melanoma.
The bubble plots of top tumor stage-associated RNA editing events (P < 0.05 and edited tumor samples ≥ 50) in tumor genes for each cancer type The X-axis shows log10P value, and the Y-axis represents the editing events and their host tumor genes. The size of each dot denotes the absolute correlation coefficients. The red and green dots represent the RNA editing events showing positive or negative relationships with tumor stages.
The volcano plots for tumor survival-related RNA editing events (P < 0.05 and edited tumor samples ≥ 50) If HR is smaller than 1.0, the X-axis denotes the value of HR−1. Otherwise, the X-axis denotes the value of (HR−1)/HR. The Y-axis represents −log10P value. The genes shown in this figure are tumor-related genes with tumor survival-related RNA editing events. HR, hazard ratio; LAML, acute myeloid leukemia; LGG, brain lower grade glioma; SARC, sarcoma; THYM, thymoma.
The bubble plots of top tumor survival-related RNA editing events (P < 0.05 and edited tumor samples ≥ 50) in tumor genes for each cancer type The X-axis shows log10P value, and the Y-axis represents the editing events and their host tumor genes. The size of each dot denotes −log10P value. The red and green dots represent the RNA editing events showing high (HR > 1) or low (HR < 1) survival risks for cancer patients.
The potential of CAediting_543208 (IGFBP3) in KIRC cancer type This event occurred only in tumor samples (246/535) and none in controls (0/72), and was up-edited in tumor samples with higher stages. It was also related to the over-expressions of IGFBP3, which was up-regulated in tumor samples and seemed to act in an autocrine action to suppress tumor cell growth. Thus, this RNA editing event might enhance the protection functions of IGFBP3 against cancer progression and be potential as a therapeutic target.
The volcano plots for DEG-associated RNA editing events in each cancer type These events were identified according to the procedures shown in Figure 3E. The X-axis denotes log2 FC value between RNA-edited and non-edited tumor samples, and the Y-axis represents −log10P value. The genes shown in this figure are tumor-related genes with DEG-associated RNA editing events.
The bubble plots of top DEG-associated RNA editing events in tumor genes for each cancer type The X-axis shows log10P value, and the Y-axis represents the editing events and their host tumor genes. The size of each dot denotes absolute log2 FC value between RNA-edited and non-edited tumor samples. The red and green dots represent the RNA editing events positively or negatively associated with the expressions of their host genes.
An example (Chr4:157360142, CAediting_390714) showing the effects of RNA editing on gene expressions The frequencies of this editing event were positively associated with the expressions of its host gene (GRIA2) in PCPG. Since this editing event led to the significantly higher expressions of its host gene, which was also up-regulated in tumor samples, CAediting_390714 may be a pathological biomarker for the PCPG cancer type. It was also supported by 146 (183) edited tumor samples and 0 (3) edited controls in this cancer type.
The enriched KEGG pathways of the overlapped DEGs for each cancer type (P < 0.05 and Q < 0.2) The pathways colored with brown are tumor-related pathways. KEGG, Kyoto Encyclopedia of Genes and Genomes.
An example showing the effects of RNA editing on miRNA regulations An RNA editing event in Chr7:5495852 (CAediting_524911) of miR589-3p altered the miRNA binding target from original DLEU1 to LEFTY1. Due to the lost regulation of miR-589-3p, DLEU1 was highly expressed in the RNA-edited TGCT tumor samples. On the other hand, LEFTY1 was down-regulated because of the gained interactions with miR-589-3p.
The possible validation of two RNA editing candidates in CCLE A. The CAediting_1478179 and CAediting_543208 were associated with its host genes in TCGA-LUSC. B. These associations were also discovered in CCLE. CCLE, Cancer Cell Line Encyclopedia.
A potential A-to-I RNA editing biomarker and its roles in cancer metastasis A. The CAediting_279186 was abnormally edited in LUSC, and also associated with RHOA expressions. B. We tested the roles of this RNA editing event between primary and lymph node metastasis lung squamous cell carcinoma samples from CCLE. C. We also validated the roles of this RNA editing between primary and metastasis lung cancer samples from MET500.
An example showing the co-effects of three RNA editing events on the expressions of APOL1 in the KIRC cancer type A. The linear regression formula to estimate the co-effects of multiple RNA editing events on gene expressions. Ck represents the contributions of the m(k-th) RNA editing event to the expression of i-th gene. B. LASSO process to select optimal parameter λ (left) and corresponding coefficients (right) for the contributions of three RNA editing events to APOL1 expressions. C. The detailed contribution coefficients are shown in this panel. LASSO, least absolute shrinkage and selection operator.
The correlations of RNA editing frequency with ADAR expression The frequencies of 9702, 2476, and 1919 RNA editing events were significantly associated with the expressions of ADAR1, ADAR2, and ADAR3, respectively (P < 0.05 and R > 0.3 for Pearson method). In addition, there are also 28,062 RNA editing events (P ≥ 0.05 for all three ADARs with more than 50 edited samples) which were possibly regulated by genetic variations, splicing efficiency, RNA binding proteins, or some other mechanisms. ADAR, adenosine deaminase action RNA.
The potentially regulatory effects of genetic variants on RNA editing A. The associations between rs2003814 and CAediting_1478179. B. The associations between rs9607326 and CAediting_1478179. C. The associations between rs9607325 and CAediting_1478179.
The distributions of A-to-I RNA editing events A. The distributions of RNA editing events in different types of genes. B. The distributions of RNA editing events in diverse regions. C. The distributions of RNA editing events in repeats. lincRNA, long intergenic non-coding RNA; ncRNA, non-coding RNA.
The effects of RNA editing on secondary structure A. An A-to-I RNA editing event will significantly reduce MFE value to stabilize RNA structure. B. 89.32% RNA transcripts were stabilized by one or multiple RNA editing events. MFE, minimum free energy.
The comparisons of RNA editing events detected by different pipelines For comparison, we converted the genome coordinates of RNA editing events detected in previous study (PMID: 26439496) from GRCh37/hg19 to GRCh38/hg38.
The statistical results for RNA editing related cancer mechanisms
The tumor-related KEGG pathways
The potentially functional A-to-I RNA editing events
Data Availability Statement
All the analysis results involved in this study are available at https://ccsm.uth.edu/CAeditome/.