Abstract
Bacterial strains isolated from attine ants showed activity against the insect specialized fungal pathogen Escovopsis and also against the human protozoan parasite Leishmania donovani. The bioassay guided fractionation of extracts from cultures of Streptomyces sp. ICBG292, isolated from the exoskeleton of Cyphomyrmex workers, led to the isolation of Mer-A2026B (1), piericidin-A1 (2) and nigericin (3). Nigericin (3) presented high activity against intracellular amastigotes of L. donovani (IC50 0.129 ± 0.008 μM). Streptomyces puniceus ICBG378, isolated from workers of Acromyrmex rugosus rugosus, produced dinactin (4) with potent anti-L. donovani activity against intracellular amastigotes (IC50 0.018 ± 0.003 μM). Compounds 3 and 4 showed good selectivity indexes, 88.91 and 656.11 respectively, and were more active than positive control, miltefosine. Compounds 1–4 were also active against some Escovopsis strains. Compounds 1 and 2 were also produced by Streptomyces sp. ICBG233, isolated from workers of Atta sexdens, and detected in ants’ extracts by mass spectrometry, suggesting they are produced in the natural environment as defensive compounds involved in the symbiotic interaction.
Author summary
Visceral leishmaniasis, caused by Leishmania infantum and L. donovani, is characterized by high rate mortality worldwide. Current treatments for this disease suffer from toxicity, variable efficacy, requirements for parenteral administration and length of treatment regimens. New chemical entities and development of new drugs are important to overcome the impact of this protozoan disease. Actinobacterial strains, such as Streptomyces, have been a source of most naturally derived antibiotics, as well as anticancer, anthelmintic, and antifungal drugs. These microorganisms also produce small molecules important in symbiotic interactions with insects, such as fungus-growing ants, fungus-growing termites, beetles and wasps against pathogens. Several novel compounds have been reported from these microorganisms with promising biological activities. In this work we show an interesting ecologic approach for drug discovery that also shows promise for the identification of antileishmanial natural products from fungus-growing ant ecosystem. Two compounds isolated from Streptomyces strains showed potent activity against L. donovani, higher than the positive control (miltefosine) with high selectivity indexes.
Introduction
Leishmaniasis is designated as Neglected Tropical Diseases (NTDs) by the World Health Organization (WHO). The visceral leishmaniasis is the most serious clinical form, produced by two Leishmania species, L. infantum and L. donovani [1]. There are between 50–90 thousands new cases and around 20–30 thousands deaths each year due to this form of leishmaniasis [2]. The treatment of leishmaniasis is still incomplete, since available drugs are toxic and expensive, have bioavailability issues, and need to overcome parasite resistance [3]. Miltefosine, originally launched as anticancer agent [4], was the only drug approved against leishmaniasis between 1981 and 2014 [5].
Prospecting understudied sources of natural products can contribute to the discovery of new antiprotozoal pharmacophores. Streptomyces associated with insects have recently emerged as a prolific and underexplored source of antimicrobials [6]. In the quadripartite symbiosis in the fungus-growing ant ecosystem between three mutualists (Attine ant, fungal garden and symbiotic actinomycetes) and one parasite (specialized pathogenic fungus Escovopsis sp.), some interspecies interactions are mediated by small molecules [7]. The ant associated actinobacteria produce secondary metabolites to inhibit the pathogen (Escovopsis sp.) but not the crop fungus (phylum Basidiomycota) [8]. This specific ecological function can guide the discovery of natural products potentially active against human pathogens [8]. Indeed, interesting bacterial-derived natural products have been reported with a wide spectrum of biological activities such as dentigerumycin [9], 9-methoxyrebeccamycin [10] and selvamicin [11].
In an ongoing International Cooperative Biodiversity Group (ICBG) initiative [12], we have isolated several actinobacteria strains from the exoskeleton of fungus-growing ants to prospect for antifungal and antiprotozoal compounds. There are some examples of compounds presenting both antifungal and antiprotozoal activities, such as azoles [13] and amphotericin B [14]. Therefore, bacterial symbionts of attine ants represent an underexplored ecosystem to search for antiprotozoal natural products based on their antifungal activity against Escovopsis in their niches.
Materials and methods
General experimental procedures
RP HPLC was performed using a Shimadzu Prominence HPLC system and a Phenomenex Luna C6-Phenyl column (5μm, 250 x 10 mm). The mass spectrometry data for 2 and 3 were acquired with a Bruker MaXis Quadrupole Time-of-Flight MS coupled to a Waters Acquity UPLC system operated by Bruker Hystar software, and for 1 and 4 with an Accela UHPLC (Thermo Scientific, USA) apparatus with an 80 Hz photodiode array detector (PDA) coupled to a Q-Exactive Plus Orbitrap mass analyzer (Thermo Scientific, USA). NMR spectra of 1–4 were obtained in CDCl3 with a Varian Unity-Inova 500 MHz spectrometer. The LC-HRESIMS and MS/MS spectra of organic extracts of Atta sexdens workers were acquired with a UPLC (Shimadzu) coupled to a micrOTOF II mass spectrometer (Bruker Daltonics).
Insect collection and isolation of actinobacteria
About 1–10 grams of the fungal gardens of the attine ants’ colony were collected. Five ants from each colony were selected for isolation of actinobacteria. Ants were identified at genus level using genera identification keys [15–17]. Specialists in taxonomy identified respective species. Ten actinobacterial strains were isolated from Acromyrmex rugosus rugosus worker ants, eight strains were isolated from Cyphomyrmex workers and twelve strains from Atta sexdens soldier ants. The bacterium Streptomyces puniceus AB10 (strain ICBG378) was isolated from A. rugosus rugosus ants collected at USP-Ribeirão Preto campus as previously described in Ortega et al. [18]. The bacterium Streptomyces sp. ICBG292 was isolated from the exoskeleton of Cyphomyrmex workers; and Streptomyces sp. ICBG233 from exoskeleton of A. sexdens workers. Cyphomyrmex and A. sexdens ants were collected in October of 2015 at the campus of the USP-Ribeirão Preto, as part of the ICBG-Brazil project [12]. Permits for collection of biological samples and research on genetic resources were issued by SISBIO (authorization 46555–6) and CNPq (010936/2014-9).
Ants collected were washed with 500 μL of sterile deionized water, vortexed for 30 s and then plated on chitin medium supplemented with the antifungals nystatin and cycloheximide (per liter: 4g chitin, 0.7g K2HPO4, 0.3g KH2PO4, 0.5g MgSO4·5H2O, 0.01g FeSO4·7H2O, 0.01g ZnSO4·7H2O, 0.01g MnCl2·4H2O, 20g of agar, 0.04 g/L nystatin, and 0.05 g/L cycloheximide). After two weeks of growth at 28°C, bacterial colonies were subcultured onto International Streptomyces Project Medium 2 (ISP-2) agar with antifungals (0.04 g/L nystatin, and 0.05 g/L cycloheximide) [19].
Identification of actinobacteria
The DNA extraction procedure was modified from Kumar et al. [20], in which the pellet was washed in 500 μL of 10.3% sucrose, centrifuged for 1 min at 10,000 g and the supernatant discarded. Then 450 μL of TSE + lysozyme were added and incubated for 20–30 min at 37°C. After, 13 μL of proteinase K was added and incubated for another 15 min at 55°C and then 250 μL of 2% SDS, gently mixed until formation of a clear solution. Then 300 μL of phenol: chloroform pH 8.0 were added and mixed and centrifuged for 10 min at 4°C. The supernatant was transferred to another tube and 60 μL of 3M NaOAc, pH 6.0 + 700 μL of isopropanol was added. The contents were mixed until "white strings" appeared and then centrifuged for 1 min to 10,000 g, and the supernatant discarded. The pellet was washed with 70% ethanol and centrifuged again at 10,000 g for 1 min. After being left overnight to completely dry the ethanol, the DNA was resuspended in 30 μL of deionized H2O.
PCR amplification of the 16S rRNA gene of actinobacteria was performed using two primers: 27F (5'-AGAGTTTGATCMTGGCT-3') and 1492R (5'-TACGGYTACCTTGTTACGACTT-3') [21]. The EconoTac DNA Polymerase Kit (Lucigen, USA) was used and the final reaction volume of 15 μL contained: 8 μL Econotaq, 0.5 μL of each primer 27F and 1492R, 0.5 μL DMSO, 4.5 μL Deionized H2O and 1 μL DNA (10ng/μL). Amplification followed the following profile: an initial denaturation step at 94°C for 3 min followed by 32 cycles of amplification of 94°C for 30s, 60°C for 30s and 72°C for 2 minutes and a final extension step of 72°C for 5 min. The PCR product was detected by agarose gel electrophoresis and visualized by ultraviolet (UV) fluorescence after staining with ethidium bromide.
The primers 27F and 1492R were used again for the sequencing of the 16S rRNA gene. The sequencing reaction of the PCR products contained: 1.5 μL 5X buffer, 1 μL primer (10 μM), 1 μL BigDye 3.1 (Applied Biosystems), 0.5 μL DMSO, 1 μL PCR product DNA and deionized water to make up the total volume of 10 μL. The program used consisted of 95°C for 3 min, followed by 35 cycles of 96°C for 10s, 58°C for 3 min and a final extent of 72°C for 7 min. The sequencing reaction was purified with the Axyprep Mag Dyeclean purification kit (Axygen) in which 5 μL of magnetic beads solution and 31 μL of 85% ethanol were added for each reaction. The tubes were placed on a magnetic plate for 3 min and then the liquid was removed. 100 μL of 85% ethanol was added for 30s and then the liquid was discarded. 100 μL of 85% ethanol was added again for 30s and after discarded. The liquid was removed as much as possible with a pipette and left overnight to completely dry the ethanol. The DNA was resuspended in 25 μL of deionized H2O.
Sequencing was performed at the Center for Genetics and Biotechnology at the University of Wisconsin—Madison (Biotech Center, UW—Madison, WI, USA). The sequences were edited and used for assembly of the contigs in the SecMan Pro Software (DNASTAR). Contigs were used to search for homologous sequences in the NCBI—GenBank (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and Eztaxon (http://www.ezbiocloud.net/eztaxon/identify). The sequences are deposited at NCBI GenBank under Accession numbers: MK118901 (ICBG233) and MK118902 (ICBG292).
In vitro evaluation of natural products on L. donovani promastigotes, intracellular amastigotes and human macrophages
Leishmania donovani axenic cultures (strain MHOM/ET/67/HU3) were maintained in M199 medium (pH 7.4) supplemented with 10% heat-inactivated fetal calf serum (FCS) and grown at 28°C [22]. Human leukemia cells (THP-1 cell line) were maintained in RPMI-1640 (FCS 10%) and grown at 37°C and 5% CO2. Stock solutions of compounds 1–4 were prepared in 100% DMSO at 10 mM and tested in 2-fold serial dilutions (10 concentrations) in 96-well flat-bottom microtiter plates.
For the promastigote assay, L. donovani cells from axenic cultures in logarithmic growth were seeded at 1 x 105/well (M199, 80 μL) and compounds were added in serial dilutions (20 μL). All plates included negative controls (100% parasite growth) and miltefosine as a positive control. After 72 hours of incubation at 28°C, 10 μL of Alamar Blue (12.5 mg resazurin/100 mL distilled water) [23] was added to each well and then the plates were incubated for 3 hours. This indicator of cell viability permeates into viable parasites, where it is reduced by NADPH and NADH enzymes to the highly fluorescent compound resorufin [24]. Following incubation, the plates were read with a microplate fluorometer under an excitation wave length of 536 nm and an emission wave length of 588 nm. If the test compound is inactive against the L. donovani, parasite remains viable and it is able to convert resazurin into resorufin, resulting in fluorescence emission. If the test compound is active against L. donovani, the number of viable parasites is reduced, thus resulting a decrease in fluorescence [25]. Growth inhibition was expressed as a percentage of the fluorescence of the negative control wells. IC50 values were determined using SigmaPlot. Dose-response curves were fitted using log (inhibitor concentration) vs. normalized response (between 0% and 100%) with variable slope, and IC50 values were automatically calculated.
In the intracellular amastigote assay, THP-1 cells were seeded at 2 × 104/well (RPMI-1640, 100 μL) with phorbol 12-myristate 13-acetate (PMA) at 20 ng/mL for differentiation into macrophages. After incubation for 72 hours (5% CO2, 37°C), medium was aspirated and late-stage promastigotes were added (2 × 105/well, 100 μL). After 24 hours of incubation, medium was aspirated to clear extracellular parasites, compounds were added in serial dilutions (100 μL) and the plates were incubated for 120 hours. All plates included negative controls (100% parasite growth) and miltefosine as a positive control. Following incubation, medium was removed and the cells were fixed in methanol and stained with Giemsa. The average number of intracellular amastigotes per THP-1 cell was determined using an inverted microscope and a cell counter [26]. Growth inhibition was expressed as a percentage of the average number of amastigotes per macrophage in the negative control wells. IC50 values were determined as described above for the promastigote assay.
For the selectivity assay, THP-1 cells were seeded at 2 × 104/well (RPMI-1640, 100 μL) with PMA at 20 ng/mL for differentiation into macrophages [27]. After incubation for 72 hours (5% CO2, 37°C), medium was aspirated, compounds were added in serial dilutions (100 μL) and the plates were incubated for 120 h. All plates included negative controls and doxorubicin as a positive control. Following incubation, 10 μL of Alamar Blue was added to each well and then the plates were incubated for 3 hours. Next, the plates were read with a microplate fluorometer under an excitation wave length of 536 nm and an emission wave length of 588 nm. Growth inhibition was expressed as a percentage of the fluorescence of the negative control wells. IC50 values were determined as described above for the promastigote and amastigote assays.
Isolation of compounds 1–3
Seed cultures of Streptomyces sp. ICBG292 were initially grown in 40 mL of ISP-2 (4 tubes of 25 × 150 mm) in a shaker for 7 days at 28°C and 200 rpm. The bacterium was inoculated into ISP-2 broth (4 g yeast extract, 10 g malt extract, and 4 g glucose per liter) in a Fernbach flask (1 L of medium in a flask of 2.8 L + 70 g of HP20) for 7 days at 28°C and 200 rpm. The HP20 and cells were filtered and washed with water and extracted with acetone (2 L). The organic solvent was filtered and dried under vacuum. A liquid-liquid partition using ethyl acetate/water was carried out, the organic phase was separated and dried to give the crude organic extract (271.7 mg). The extract was purified by SPE-C18 (55 μm, 1 g) using the following gradient: 10 mL (20% MeOH-H2O, A1: 55.1 mg); 10 mL (40% MeOH-H2O, A2: 23.5 mg); 10 mL (60% MeOH-H2O, A3: 14.5 mg); 10 mL (80% MeOH-H2O, A4: 47.2 mg); and 10 mL (100% MeOH, A5: 85.5 mg). Fractions A4 and A5 were active against Escovopsis (S13 Fig), so they were combined and further purified by semi-preparative HPLC using the column C6-Phenyl (5μm, 250 x 10 mm) and the following gradient at 4 mL/min: 1–20 min, linear gradient from 70% MeOH-H2O to 100% MeOH; 20–25 min, isocratic flow of 100% MeOH; 25–25.5 min, linear gradient from 100% MeOH to 70% MeOH-H2O; 25.5–30.5 min, isocratic flow of 70% MeOH-H2O to give 13 fractions [A5.1 (2.5 mg); A5.2 (5.0 mg); A5.3 (3.1 mg); A5.4 (2.6 mg); A5.5 (3.1 mg); A5.6 (15.5 mg), A5.7 (10.9 mg), A5.8 (2.2 mg), A5.9 (14.3 mg), A5.10 (1.6 mg), A5.11 (0.9 mg), A5.12 (1.2 mg), A5.13 (4.3 mg)]. Fractions A5.2, A5.6 and A5.9 were identified by NMR and HRESIMS as antibiotics Mer-A2026B (1), piericidin-A1 (2) and nigericin (3), respectively (S1–S6 Figs). Purity of compounds was measured by HPLC as 99% for compound 1, 97% for compound 2 and 93% for compound 3.
Isolation of compound 4
Seed culture of S. puniceus ICBG378 was initially grown in 10 mL of ISP-2 (25 × 150 mm tube) and was mounted into a shaker for 2 days at 28°C and 200 rpm. The bacterium was inoculated in broth A-medium (20 g soluble starch, 10 g glucose, 5 g peptone, 5 g yeast extract, 5 g CaCO3 per liter) in a baffled Erlenmeyer flask [2 x (100 mL of medium in a flask of 500 mL + 4 mL of seed culture + 7 g of HP20)] for 7 days at 28°C and 200 rpm. The HP20 was filtered and washed with distilled water and acetone. The organic solvent was filtered and dried under vacuum to give the crude extract (235.68 mg), which was purified by SPE-ENV+ (55 μm, 1 g) using the following gradient: 10 mL (25% MeOH-H2O, B1: 50.1 mg); 10 mL (50% MeOH-H2O, B2: 20.5 mg); 10 mL (75% MeOH-H2O, B3: 41.3 mg); 10 mL (100% MeOH, B4: 102.5 mg). Fractions B3 and B4 were active against Escovopsis (S13 Fig). They were mixed and purified by SPE-Si (55 μm, 500 mg) with the gradient: 8 mL [100% Hexane, B4.1: 32.6 mg], 8 mL [Hexane:EtOAc (8:2), B4.2: 57.0 mg], 8 mL [Hexane:EtOAc (6:4), B4.3: 8.9 mg], [Hexane:EtOAc (4:6), B4.4: 6.7 mg], 8 mL [Hexane:EtOAc (2:8), B4.5: 3.5 mg], 8 mL [100% EtOAc, B4.6: 2.9 mg], and 8 mL [100% Methanol, B4.7: 15.3 mg]. The fraction B4.2 was identified by NMR and HRESIMS as dinactin (4) with 91% purity as measured by HPLC (S7and S8 Figs).
Antagonist bioassay of bacterial strains and compounds against fungi
Each bacterium-fungus and compound-fungal challenge was replicated two times on ISP-2 agar. Bacteria strains were initially screened against Escovopsis sp. ICBG1251. Bacteria were placed in the center of ISP-2 agar Petri dishes and grown alone during 7 days; fungal strains were then point-inoculated near the edge of the culture (microbial strains distant from each other around 3 cm). Two microliters of compounds (100 μg) were placed in the center of Petri dishes and fungal strains were then point-inoculated near the edge of the plate. The positive control used was the miconazole. Challenges were monitored each 7 days and inhibition zone was measure after 21 days [8]. Four different fungal strains were used for testing the pure compounds: Escovopsis sp. ICBG711 (from Trachymyrmex colony), Escovopsis sp. ICBG740 (from Acromyrmex colony), Escovopsis sp. ICBG1251 (from Atta colony) and Trichoderma sp. ICBG1100 (from attine colony).
Identification of compounds 1 and 2 from Atta sexdens exoskeleton
Atta sexdens colonies, collected at USP-Ribeirão Preto campus, were kept under laboratorial conditions. A total of 25 A. sexdens individuals, obtained from these colonies, were mechanically cleaned using small forceps and extracted with 50 mL of methanol. The extracts were filtered and evaporated to dryness. The crude extracts were evaluated for the presence or absence of compounds 1 and 2 by LC-HRESIMS, as described at general procedures.
Results and discussion
Ten bacterial strains were isolated from A. rugosus rugosus ants, eight strains were recovered from Cyphomyrmex ants and twelve from A. sexdens. All 30 bacterial strains were challenged in antagonism assays against Escovopsis sp., the specialized pathogenic fungus of Attine ants, and bioactive strains were identified through 16S rRNA sequencing. Streptomyces puniceus ICBG378 from A. rugosus rugosus, Streptomyces sp. ICBG292 from Cyphomyrmex sp., and Streptomyces sp. ICBG233 from A. sexdens showed high inhibition of Escovopsis, and were selected for scale up culturing and antiprotozoal assays. Crude extracts and fractions of cultures of the three selected Streptomyces strains inhibited the growth of L. donovani promastigotes (inhibition higher than 90%). Therefore, they were selected for the isolation and characterization of biologically active natural products.
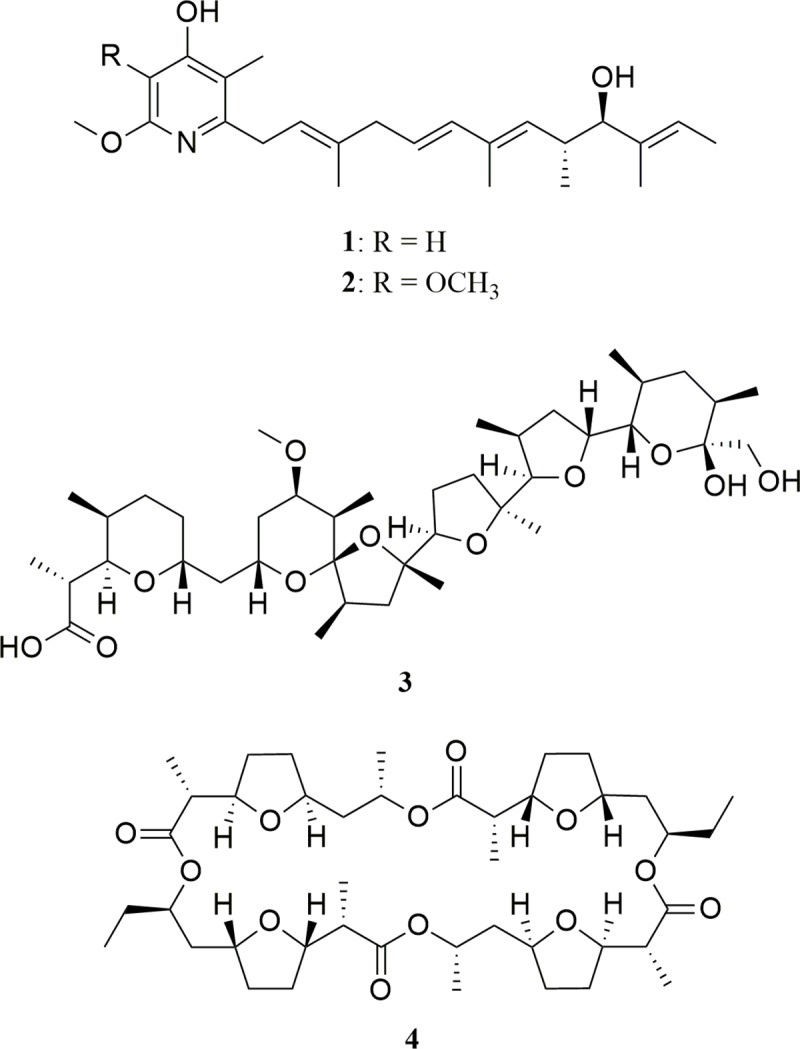
The fractionation of extracts was guided by the antifungal assay against Escovopsis (S1 Fig) and led to the isolation of the known antibiotics mer-A2026B [28] (1), piericidin-A1 [29] (2), nigericin [30,31] (3), produced by Streptomyces sp. ICBG292 (Fig 1); and dinactin [32] (4), produced by S. puniceus ICBG378. Compounds 1 and 2 were also isolated from Streptomyces sp. ICBG233, associated with A. sexdens ants. Structures were established on the basis of NMR and HRESIMS data and comparison with literature (S2–S14 Figs).
Fig 1. Compounds identified from bacteria Streptomyces sp. ICBG292 (1–3), Streptomyces sp. ICBG233 (1, 2), and S. puniceus ICBG378 (4).
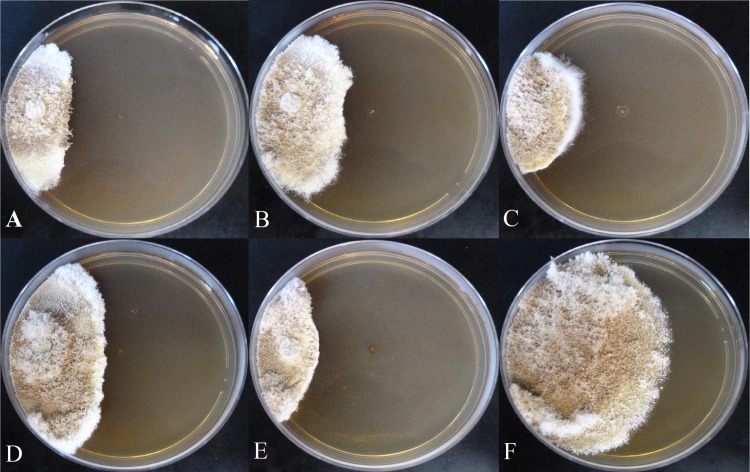
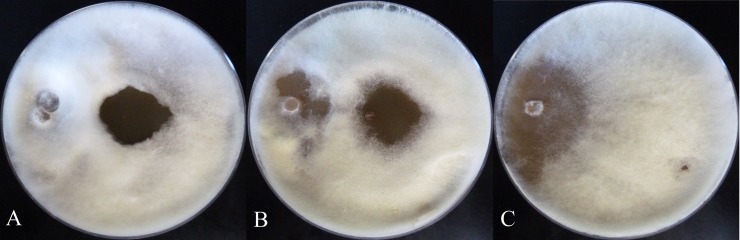
Compounds 1–4 were active against Escovopsis sp. ICBG740 (Fig 2). Compound 1 showed higher antagonist activity against four different Escovopsis strains compared to compounds 2–4 (Fig 2and S15–S17 Figs), with inhibition zone similar to the positive control (miconazole). Compound 1 was also active against the fungus Trichoderma sp. (Fig 3).
Fig 2. ICBG741 (21 days of growth at 28°C).
Antagonist activity of compounds (100 μg) against Escovopsis sp. A) Mer-A2026B (1), B) piericidin-A1 (2), C) nigericin (3), D) dinactin (4), E) miconazole, and F) negative control.
Fig 3. Antagonist activity of compounds (100 μg) against Trichoderma sp ICBG1100 (21 days of growth at 28°C).
A) Mer-A2026B (1), B) Miconazole, C) Negative control.
All compounds were active against L. donovani promastigotes while compounds 1, 2 and 4 were also active against intracellular amastigotes (Table 1). This is the first report of the antileishmanial activity of antibiotics Mer-A2026B (1), piericidin-A1 (2) and dinactin (4). L. donovani lives in the sandfly gut as promastigotes. Promastigotes are the infective stage of Leishmania sp., being transmitted to humans via the bite of sandflies. Skin macrophages phagocyte the promastigotes, where the promastigotes differentiate into amastigote form. Intracellular amastigotes reproduce within the macrophages, eventually rupturing the host cell to infect other surrounding macrophages [33]. In addition to being involved in different stages of the life cycle of the parasite, promastigotes and amastigotes differ morphologically. Promastigotes are flagellated elongated cells, while amastigotes are rounded non-flagellated cells [26]. Compounds 3 and 4 were more active against both L. donovani forms than the positive control miltefosine (Table 1). Although intracellular amastigotes are the clinically relevant form, assessing the activity against both parasite stages can provide important information for further studies on the mechanism of action of these compounds. These activity data can be useful to investigate which biochemical pathways are modulated and understand the role played by the respective molecular targets in each stage of the parasite life cycle. The selectivity index, which is the ratio between the activity against THP-1 macrophages and intracellular amastigotes, indicates whether the compounds are selective for L. donovani over the human host cells. The probability of a compound to elicit cytotoxic effects in the human host decreases as the selectivity index increases. Therefore, the selectivity index is an important safety metric, and was assessed for compounds 3 and 4. The selectivity indexes of 3 and 4 were 88.91 and 656.11, respectively, suggesting their safety.
Table 1. Activity of compounds 1–4 on L. donovani intracellular amastigotes, against promastigotes and THP-1.
| Compounds | IC50 (μM) Intracellular amastigotes | IC50 (μM) Promastigotes | CC50 (μM) THP-1a | Selectivity Indexb |
|---|---|---|---|---|
| 1 | 49.85 ± 7.01 | 35.86 ± 1.83 | > 64 | — |
| 2 | > 64 | 38.41 ± 4.63 | > 64 | — |
| 3 | 0.129 ± 0.008 | 0.284 ± 0.072 | 11.47 ± 0.68 | 88.91 |
| 4 | 0.018 ± 0.003 | 0.032 ± 0.005 | 11.81 ± 1.57 | 656.11 |
| Doxorubicin | — | — | 0.571 ± 0.068 | — |
| Miltefosine | 5.80 ± 0.59 | 4.74 ± 0.25 | — | — |
Data are shown as mean ± SD (n = 2 biological replicates)
aTHP-1 human leukemia macrophages (host cells of L. donovani)
bSelectivity index = CC50 THP-1/IC50 intracellular amastigotes
A potent vasodilating activity has been reported for mer-A2026B (1) [34]; and insecticidal, antimicrobial and cytotoxic activities for piericidin-A1 (2) [35–37]; while strong antibacterial and anticancer activities have been found for nigericin (3) and dinactin (4) [38–40]. The high activities and good selectivity indexes obtained for nigericin (3) and dinactin (4) in our experiments (Table 1) are in agreement with previous data for nigericin monosodium salt and nonactin, an analogue of 4, using ex-vivo splenic explant culture system from hamsters infected with L. donovani [41].
Compounds 3 and 4 are considered ionophores that reversibly bind and transport ions across biological membranes [42]. Nigericin (3) has been shown to move sodium and potassium ions through membranes [43]. When bound to a cation, nigericin loses a proton and generates an uncharged species that can permeate into cell membranes, acting as a carrier. The molecule can also permeate into membranes as a protonated noncomplexed molecule. Nigericin can promote an exchange of K+ for H+ that results in the modification of the ion gradient across the membranes involved in the energetic metabolism [44]. Dinactin (4) is one member of the family of macrotetrolide nactins with ability to selectively complex a wide variety of cations [45]. Few ionophore compounds have been described to inhibit L. donovani. One example is the ionophore A23187 that binds Ca2+ and kills intracellular Leishmania in the presence of lipopolysaccharide (LPS), mediated by generation of L-arginine-dependent nitrogen oxidation products [46]. Another ionophore, named calcimycin, has been described to kill Leishmania promastigotes by activating parasite nitric oxide synthase [47]. The Leishmania cell death is accompanied by the loss of mitochondrial polarization and plasma membrane integrity and can be blocked by specific inhibitors of constitutive Ca2+/calmodulin-dependent nitric oxide synthase [47]. The most recognized mechanism of action of miltefosine against L. donovani is the inhibition of phospholipid synthesis and cytochrome c oxidase, but recently another mechanism has been described based in the abrupt increase in the intracellular Ca2+ concentration in the L. donovani [48], a similar property of ionophores.
L. donovani lives in the sandfly gut as promastigotes and in mammalian macrophages as amastigotes [49]. This protozoan extrudes protons through H+-ATPase to regulate intracellular pH and to facilitate nutrient uptake [49]. This proton extrusion is enhanced by the addition of K+ [49]. This could be one mechanism by which nigericin controls the growth of Leishmania parasite. The mechanisms of action of nigericin (3) and dinactin (4) against L. donovani have not been described.
Compounds 1 and 2 showed drug-like properties according to several rules such as Lipinski and Veber filters [50,51], while 3 and 4 exceed the ideal molecular weight and number of hydrogen-bond acceptors (HBA) (Table 2). The computational predictions were run using SwissADME [52] and Stardrop (Optibrium) [53].
Table 2. Molecular properties of compound 1–4.
| Code | MW | LogP | hERGpIC50 | HIA | HBD | HBA | TPSA | nrotb | Drug-likeness | PAINS alert |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 385.54 | 4.10 | 5.93 | + | 2 | 4 | 62.58 | 9 | yes | 0 |
| 2 | 415.56 | 3.93 | 5.93 | + | 2 | 5 | 71.81 | 10 | yes | 0 |
| 3 | 724.96 | 3.77 | 3.96 | - | 3 | 11 | 142.37 | 9 | no | 0 |
| 4 | 764.98 | 4.70 | 4.43 | + | 0 | 12 | 142.10 | 2 | no | 0 |
| Desired Value | ≤ 500 | < 5 | < 6.3 | + | ≤ 5 | ≤ 10 | ≤ 140 Å | ≤ 10 |
LogP: octanol/water partition coefficient; hERG pIC50: -logIC50 on human ether-a-go-go-related gene potassium ion channels; HIA: human intestinal absorption; HBD: hydrogen-bond donors; HBA: hydrogen-bond acceptors; TPSA: topological surface area; nrotb: number of rotatable bonds; PAINS: pan-assay interference compounds. Drug-likeness according to Lipinski and Veber filters.
Lipinski’s rule of five states that compounds showing more than 5 hydrogen-bond donors, 10 hydrogen-bond acceptors, molecular weight greater than 500 and LogP greater than 5, are likely to show poor gastrointestinal absorption [50]. However, several natural products that do not comply with Lipinski´s rules have been approved as drugs, such as paclitaxel, rapamycin, cyclosporine A, and others. In general, natural products are considered as exceptions to Lipinski´s rules. However, the properties LogP and hydrogen-bond donors are very important for predicting bioavailability. A possible explanation is that nature can maintain low hydrophobicity and intermolecular hydrogen-bond donating potential when it needs to produce active compounds with high molecular weight and rotatable bonds; and natural products could also take advantage of active transport mechanisms since they contain biosynthetic moieties that resemble endogenous metabolites [54]. So, dinactin (4) could be an interesting compound for further pharmacological studies in the treatment of leishmaniasis based on the high selectivity index against L. donovani and on LogP and HBD values that comply with Lipinski´s rules. Furthermore, given their remarkable in vitro activity, compounds 3 and 4 are suitable starting points for molecular optimization aiming to pursue molecules that fit into the drug-like concept.
Compounds 1–4 can join the chemical cocktail used by actinobacteria to control the growth of the pathogenic fungus Escovopsis sp. and other opportunistic fungi such as Trichoderma sp. in fungus-growing ant colonies. Compounds 1 and 2 were also identified from Streptomyces sp. ICBG233 associated to workers of Atta sexdens and from the organic extract of these ants by mass spectrometry (S18–S23 Figs), confirming their production in the natural environment. Compound 2 and other piericidin derivatives together with nigericin (3) have also been reported from Candidatus Streptomyces philanthi symbiont of solitary beewolf digger wasps (Philanthus triangulum, Hymenoptera, Crabronidae) as antibiotic protectors of their larval offspring against pathogens [55,56]. Authors argue that the mixture of these antibiotics could help in the evolutionary stable defense against different pathogens [55,56], and the current identification of the same compounds in bacterial symbionts of attine ants reinforces this hypothesis.
Considering the remarkable activity against L. donovani shown by the identified compounds and that the treatment for visceral leishmaniasis suffers from several drawbacks, the results reported herein can contribute to the development of novel therapeutic agents for this NTD. Moreover, most current drug development approaches are based on high-throughput screening (HTS) of synthetic compound collections. HTS platforms can screen libraries containing thousands of molecules, whose chemical diversity are provided by methods such as combinatorial chemistry. Natural products can provide further structural diversity and novel chemotypes that differ from those obtained via combinatorial chemistry. In this context, natural products are a rich source of structural diversity that offers unique chemical matter to be used as reference for the design of novel leishmanicidal agents. Our results also validate the ecological approach of screening antifungal natural products from actinobacteria associated to attine ants as a good strategy for discovering antileishmanial compounds.
Supporting information
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Region of 10 to 46 ppm.
(PDF)
Region of 56 to 110 ppm.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
We acknowledge Claudia C. Macedo for the technical support. Diego Santana Assis (Faculty of Philosophy, Sciences and Letters of Ribeirão Preto (FFCLRP) University of São Paulo (USP) and Sandra Verza da Silva (Institute of Biosciences, State University of São Paulo “Julio de Mesquita Filho” (UNESP), Rio Claro, SP) are acknowledged for the identification of attine ants.
Data Availability
The 16S rRNA sequences of actinobacteria are deposited at NCBI GenBank under Accession numbers: MK118901 (ICBG233) and MK118902 (ICBG292). All other relevant data are within the manuscript and its Supporting Information files.
Funding Statement
We received financial support from São Paulo Research Foundation (FAPESP) grants #2013/50954-0 (MTP), #2014/14095-6 (HEO), #2016/20154-0 (HEO), #2015/01001-6 (WGPM), #2013/07600-3 (CEPID-CIBFar) and #2016/15872-1 (ALLO); Fogarty International Center, National Institutes of Health (FIC-NIH) grant U19TW009872 (TSB); Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) grant #307147/2014-2 (MTP). This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) - Finance Code 001. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Chappuis F, Sundar S, Hailu A, Ghalib H, Rijal S, Peeling RW, et al. Visceral leishmaniasis: what are the needs for diagnosis, treatment and control? Nat Rev Microbiol. 2007; 5(11):873–82. 10.1038/nrmicro1748 [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. Leishmaniasis. 2019. https://www.who.int/leishmaniasis/en/ [Google Scholar]
- 3.Croft SL, Sundar S, Fairlamb AH. Drug Resistance in Leishmaniasis. Clin Microbiol Rev. 2006; 19(1):111–26. 10.1128/CMR.19.1.111-126.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dorlo TPC, Balasegaram M, Beijnen JH, de vries PJ. Miltefosine: A review of its pharmacology and therapeutic efficacy in the treatment of leishmaniasis. J Antimicrob Chemother. 2012; 67(11): 2576–97. 10.1093/jac/dks275 [DOI] [PubMed] [Google Scholar]
- 5.Newman DJ, Cragg GM. Natural Products as Sources of New Drugs from 1981 to 2014. J Nat Prod. 2016; 79(3):629–61. 10.1021/acs.jnatprod.5b01055 [DOI] [PubMed] [Google Scholar]
- 6.Chevrette MG, Carlson CM, Ortega HE, Thomas C, Ananiev GE, Barns KJ, et al. The antimicrobial potential of Streptomyces from insect microbiomes. Nat Commun. 2019; 10(1):516 10.1038/s41467-019-08438-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schultz TR, Brady SG. Major evolutionary transitions in ant agriculture. Proc Natl Acad Sci. 2008; 105(14):5435–40. 10.1073/pnas.0711024105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Currie CR, Scottt JA, Summerbell RC, Malloch D. Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature. 1999; 398:701–4. 10.1038/19519 [DOI] [Google Scholar]
- 9.Oh DC, Poulsen M, Currie CR, Clardy J. Dentigerumycin: A bacterial mediator of an ant-fungus symbiosis. Nat Chem Biol. 2009; 5(6):391–3. 10.1038/nchembio.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Arnam EB, Ruzzini AC, Sit CS, Currie CR, Clardy J. A Rebeccamycin Analog Provides Plasmid-Encoded Niche Defense. J Am Chem Soc. 2015; 137(45):14272–4. 10.1021/jacs.5b09794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Arnam EB, Ruzzini AC, Sit CS, Horn H, Pinto-Tomás AA, Currie CR, et al. Selvamicin, an atypical antifungal polyene from two alternative genomic contexts. Proc Natl Acad Sci. 2016; 113(46):12940–5. 10.1073/pnas.1613285113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pupo MT, Currie CR, Clardy J. Microbial symbionts of insects are the focus of the first International Cooperative Biodiversity Group (ICBG) in Brazil. J Braz Chem Soc. 2017; 28(3):393–401. 10.21577/0103-5053.20160284. [DOI] [Google Scholar]
- 13.Saenz RE, Paz H, Berman JD. Efficacy of ketoconazole against Leishmania braziliensis panamensis cutaneous leishmaniasis. Am J Med. 1990; 89(2):147–55. 10.1016/0002-9343(90)90292-l [DOI] [PubMed] [Google Scholar]
- 14.New RR, Chance ML, Heath S. Antileishmanial activity of amphotericin and other antifungal agents entrapped in liposomes. J Antimicrob Chemother. 1981; 8(5):371–81. 10.1093/jac/8.5.371 [DOI] [PubMed] [Google Scholar]
- 15.Bolton B. Identification guide to the ant genera of the world 1st ed. Harvard University Press; 1994. ISBN 9780674442801. http://www.hup.harvard.edu/catalog.php?isbn=9780674442801 [Google Scholar]
- 16.Bolton B. A new general catalogue of the ants of the world 1st ed. Harvard University Press; 1995. ISBN 9780674615144. http://www.hup.harvard.edu/catalog.php?isbn=9780674615144 [Google Scholar]
- 17.Baccaro FB, Feitosa RM, Fernandez F, Fernandes IO, Izzo TJ, De Souza JLP, Solar R. Guia para os gêneros de formigas do Brasil 1st ed. Manaus: Editora INPA; 2016. ISBN 978-85-211-0152-9. https://ppbio.inpa.gov.br/sites/default/files/Livro_Formigas_2015.pdf [Google Scholar]
- 18.Ortega HE, Batista JM, Melo WGP, Clardy J, Pupo MT. Absolute configurations of griseorhodins A and C. Tetrahedron Lett. 2017; 58(50):4721–3. 10.1016/j.tetlet.2017.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poulsen M, Currie CR. Symbiont interactions in a tripartite mutualism: Exploring the presence and impact of antagonism between two fungus-growing ant mutualists. PLoS One. 2010; 5(1). 10.1371/journal.pone.0008748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar V, Bharti A, Gusain O, Bisht GS. An Improved Method for Isolation of Genomic DNA from Filamentous Actinomycetes. J Eng Technol Manag. 2010; 2(2):10–13. [Google Scholar]
- 21.Ludwig W. Nucleic acid techniques in bacterial systematics and identification. Int J Food Microbiol. 2007; 120(3):225–36. 10.1016/j.ijfoodmicro.2007.06.023 [DOI] [PubMed] [Google Scholar]
- 22.Gupta S. Visceral leishmaniasis: experimental models for drug discovery. Indian J Med Res. 2011; 133(1):27–39. http://www.ijmr.org.in/text.asp?2011/133/1/27/76700 [PMC free article] [PubMed] [Google Scholar]
- 23.Ioset JR, Brun R, Wenzler T, Kaiser M, Yardley V. Drug Screening for Kinetoplastids Diseases A Training Manual for Screening in Neglected Diseases. DNDi Pan-Asian Screen Netw. 2009. https://www.dndi.org/2009/media-centre/scientific-articles/scientific-articles-vl/drug-screening-for-kinetoplastid-diseases-a-training-manual-for-screening-in-neglected-diseases-2/ [Google Scholar]
- 24.Shimony O, Jaffe CL. Rapid fluorescent assay for screening drugs on Leishmania amastigotes. J Microbiol Methods. 2008; 75(2):196–200. 10.1016/j.mimet.2008.05.026 [DOI] [PubMed] [Google Scholar]
- 25.Mikus J, Steverding D. A simple colorimetric method to screen drug cytotoxicity against Leishmania using the dye Alamar Blue. Parasitology International. 2000; 48(3):265–269. 10.1016/S1383-5769(99)00020-3 [DOI] [PubMed] [Google Scholar]
- 26.Zulfiqar B, Shelper TB, Avery VM. Leishmaniasis drug discovery: recent progress and challenges in assay development. Drug Discovery Today. 2017; 22(10):1516–31. 10.1016/j.drudis.2017.06.004 [DOI] [PubMed] [Google Scholar]
- 27.Karwaciak I, Gorzkiewicz M, Bartosz G, Pulaski L. TLR2 activation induces antioxidant defence in human monocyte-macrophage cell line models. Oncotarget. 2017; 8(33):54243–64. 10.18632/oncotarget.17342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kominato K, Watanabe Y, Hirano S, Kioka T, Terasawa T, Yoshioka T, et al. Mer-A2026A and B, novel piericidins with vasodilating effect. I. Producing organism, fermentation, isolation and biological properties. J Antibiot (Tokyo). 1995; 48(2):99–102. 10.7164/antibiotics.48.99 [DOI] [PubMed] [Google Scholar]
- 29.Yoshida S, Yoneyama K, Shiraishi S, Watanabe A, Takahashi N. Chemical Structures of New Piericidins Produced by Streptomyces pactum. Agric Biol Chem. 1977;41(5):855–62. 10.1080/00021369.1977.10862594 [DOI] [Google Scholar]
- 30.Steinrauf LK, Pinkerton M, Chamberlin JW. The structure of nigericin. Biochem Biophys Res Commun. 1968; 33(1):29–31. 10.1016/0006-291x(68)90249-0 [DOI] [PubMed] [Google Scholar]
- 31.Seto H, Mizove K, Nakayama H, Furihata K, Otake N, Yonehara H. Studies on the ionophorous antibiotics. XX. Some empirical rules for structural elucidation of polyether antibiotics by 13C-NMR spectroscopy. J Antibiot (Tokyo). 1979; 32(3):239–43. 10.7164/antibiotics.32.239 [DOI] [PubMed] [Google Scholar]
- 32.Beck J, Gerlach H, Prelog V, Voser W. Stoffwechselprodukte von Actinomyceten. 35. Mitteilung. Über die Konstitution der Makrotetrolide Monactin, Dinactin und Trinactin. Helv Chim Acta. 1962; 45(2):620–30. http://doi.wiley.com/10.1002/hlca.19620450227 [Google Scholar]
- 33.Burza S, Croft SL, Boelaert M. Leishmaniasis. Lancet (London, England). 2018; 392(10151): 951–970. 10.1016/S0140-6736(18)31204-2 [DOI] [PubMed] [Google Scholar]
- 34.Kominato K, Watanabe Y, Hirano S, Kioka T, Terasawa T, Yoshioka T, et al. Mer-A2026A and B, novel piericidins with vasodilating effect. I. Producing organism, fermentation, isolation and biological properties. J Antibiot (Tokyo). 1995; 48(2):99–102. 10.7164/antibiotics.48.99 [DOI] [PubMed] [Google Scholar]
- 35.Schnermann MJ, Romero FA, Hwang I, Nakamaru-Ogiso E, Yagi T, Boger DL. Total Synthesis of Piericidin A1 and B1 and Key Analogues. J Am Chem Soc. 2006, 128(36):11799–807. 10.1021/ja0632862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamura S, Takahashi N, Miyamoto S, Mori R. Isolation and Physiological Activities of Piericidin A, A Natural Insecticide Produced by Streptomyces. Agric Biol Chem. 1963;27(8):576–82. 10.1080/00021369.1963.10858144 [DOI] [Google Scholar]
- 37.Urakawa A, Sasaki T, Yoshida K-I, Otani T, Lei Y, Yun W. IT-143-A and B, Novel Piericidin-group Antibiotics Produced by Streptomyces sp. J Antibiot (Tokyo). 1996;49(10):1052–5. 10.7164/antibiotics.49.1052 [DOI] [PubMed] [Google Scholar]
- 38.Wu Z, Bai L, Wang M, Shen Y. Structure–antibacterial relationship of nigericin derivatives. Chem Nat Compd. 2009; 45(3):333–7. 10.1007/s10600-009-9350-x [DOI] [Google Scholar]
- 39.Liu F, Li W, Hua S, Han Y, Xu Z, Wan D, et al. Nigericin exerts anticancer effects on human colorectal cancer cells by inhibiting Wnt/β-catenin signaling pathway. Mol Cancer Ther. 2018; 17(5):952–65. 10.1158/1535-7163.MCT-17-0906 [DOI] [PubMed] [Google Scholar]
- 40.Hussain A, Rather MA, Dar MS, Dangroo NA, Aga MA, qayum A, et al. Streptomyces puniceus strain AS13., Production, characterization and evaluation of bioactive metabolites: A new face of dinactin as an antitumor antibiotic. Microbiol Res. 2018; 207:196–202. 10.1016/j.micres.2017.12.004 [DOI] [PubMed] [Google Scholar]
- 41.Osorio Y, Travi BL, Renslo AR, Peniche AG, Melby PC. Identification of small molecule lead compounds for visceral leishmaniasis using a novel ex vivo splenic explant model system. PLoS Negl Trop Dis. 2011; 5(2): e962 10.1371/journal.pntd.0000962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bakker E, Bühlmann P, Pretsch E. Carrier-based ion-selective electrodes and bulk optodes. 1. General characteristics. Chem Rev. 1997; 97(8):3083–3132. https://pubs.acs.org/doi/10.1021/cr940394a [DOI] [PubMed] [Google Scholar]
- 43.Riddell FG, Arumugam S, Brophy PJ, Cox BG, Payne MCH, Southon TE. The Nigericin-Mediated Transport of Sodium and Potassium Ions through Phospholipid Bilayers Studied by sodium-23 and potassium-39 NMR Spectroscopy. J Am Chem Soc. 1988; 110(3):734–8. 10.1021/ja00211a012 [DOI] [Google Scholar]
- 44.Nicholls DG, Ferguson SJ, Nicholls DG, Ferguson SJ. Bioenergetics 4 Ion Transport Across Energy-Conserving Membranes. Academic Press; 2013. pp. 13–25. 10.1016/B978-0-12-388425-1.00002-6 [DOI] [Google Scholar]
- 45.Asher IM, Phillies GDJ, Kim BJ, Stanley HE. Ion complexation in nonactin, monactin, and dinactin: A Raman spectroscopic study. Biopolymers. 1977; 16(1):157–85. 10.1002/bip.1977.360160112 [DOI] [PubMed] [Google Scholar]
- 46.Buchmuller-Rouiller Y, Corradin SB, Mauel J. Macrophage activation for intracellular killing as induced by a Ca2+ ionophore. Biochem J. 1992; 284:387–92. 10.1042/bj2840387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grekov I, Pombinho AR, Kobets T, Bartůněk P, Lipoldová M. Calcium Ionophore, Calcimycin, Kills Leishmania Promastigotes by Activating Parasite Nitric Oxide Synthase. Biomed Res Int. 2017; 2017:1–6. 10.1155/2017/1309485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pinto-Martinez AK, Rodriguez-Durán J, Serrano-Martin X, Hernandez-Rodriguez V, Benaim G. Mechanism of action of miltefosine on Leishmania donovani involves the impairment of acidocalcisome function and the activation of the sphingosine-dependent plasma membrane Ca2+ channel. Antimicrob Agents Chemother. 2018; 62(1):1–10. 10.1128/AAC.01614-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang S, Anderson SA, Winget GD, Mukkada AJ. Plasma membrane K+/H+-ATPase From leishmania donovani. J Cell Physiol. 1994; 159(1):60–6. 10.1002/jcp.1041590109 [DOI] [PubMed] [Google Scholar]
- 50.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001; 46(1–3):3–26. 10.1016/S0169-409X(00)00129-0 [DOI] [PubMed] [Google Scholar]
- 51.Veber DF, Johnson SR, Cheng H-Y, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem. 2002; 45(12):2615–23. 10.1021/jm020017n [DOI] [PubMed] [Google Scholar]
- 52.Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017; 7(1):42717 10.1038/srep42717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Segall M. Advances in multiparameter optimization methods for de novo drug design. Expert Opin Drug Discov. 2014; 9(7):803–17. 10.1517/17460441.2014.913565 [DOI] [PubMed] [Google Scholar]
- 54.Ganesan A. The impact of natural products upon modern drug discovery. Curr Opin Chem Biol. 2008; 12(3):306–17. 10.1016/j.cbpa.2008.03.016 [DOI] [PubMed] [Google Scholar]
- 55.Engl T, Kroiss J, Kai M, Nechitaylo TY, Svatoš A, Kaltenpoth M. Evolutionary stability of antibiotic protection in a defensive symbiosis. Proc Natl Acad Sci. 2018; 115(9): E2020–E2029. 10.1073/pnas.1719797115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kroiss J, Kaltenpoth M, Schneider B, Schwinger MG, Hertweck C, Maddula RK, et al. Symbiotic streptomycetes provide antibiotic combination prophylaxis for wasp offspring. Nat Chem Biol. 2010; 6(4):261–3. 10.1038/nchembio.331 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Region of 10 to 46 ppm.
(PDF)
Region of 56 to 110 ppm.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
The 16S rRNA sequences of actinobacteria are deposited at NCBI GenBank under Accession numbers: MK118901 (ICBG233) and MK118902 (ICBG292). All other relevant data are within the manuscript and its Supporting Information files.